News
2024-08-30
Launch of Two New Research Projects in PBPK/PD Modeling
Ennie Tensil and Michelle Elias start Bachelor projects on losartan and glimeperide
We are excited to announce the initiation of two projects within our group, focusing on the development of Physiologically-Based Pharmacokinetic/Pharmacodynamic (PBPK/PD) models. These projects aim to deepen our understanding of drug action and variability in treatment responses, contributing significantly to personalized medicine. Michelle Elias will lead the first project, which centers on the development of a PBPK/PD model for the sulfonylurea glimepiride, a widely used medication for managing type 2 diabetes. This project aims to enhance our understanding of the intraindividual variability observed in diabetes treatment with sulfonylureas, particularly focusing on differences in hepatorenal function. The insights gained from this model could lead to more personalized and effective treatment strategies for patients with diabetes. The second project, led by Ennie Tensil, will focus on developing a PBPK/PD model for the angiotensin II receptor blocker (ARB) losartan. This medication is commonly prescribed for treating high blood pressure and protecting against stroke in high-risk individuals. The project will investigate the physiological factors influencing the pharmacokinetics and pharmacodynamics of losartan, aiming to improve our understanding of its efficacy and variability in different patient populations. Both projects represent important advancements in our research capabilities, with the potential to impact clinical practice by providing deeper insights into drug behavior and patient variability. We look forward to the outcomes these studies will bring and the contributions they will make to the field of pharmacology.
2024-08-28
Master Thesis Jonas Küttner
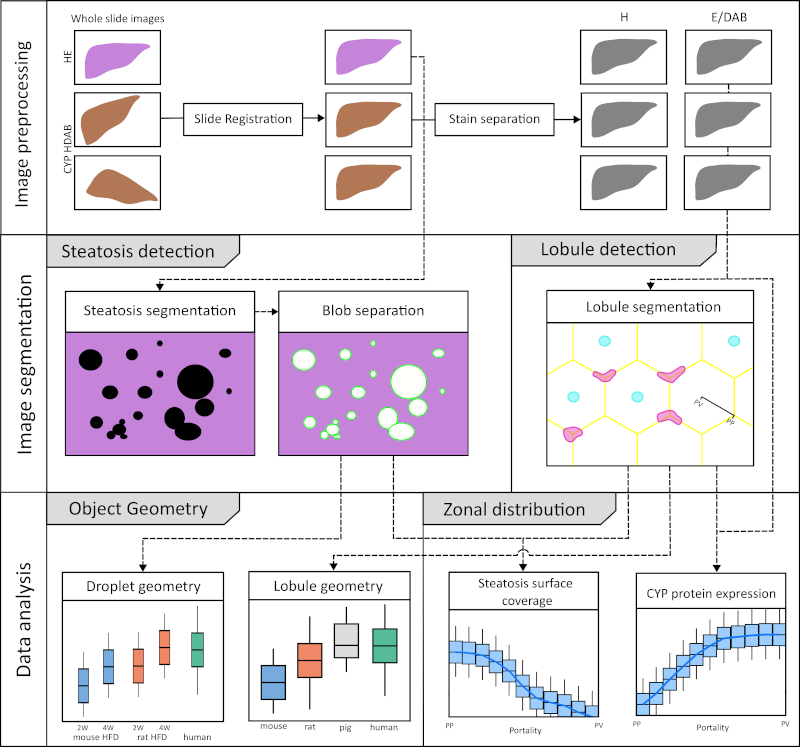
Jonas Küttner submitted his Master thesis developing a workflow for image analysis in the liver
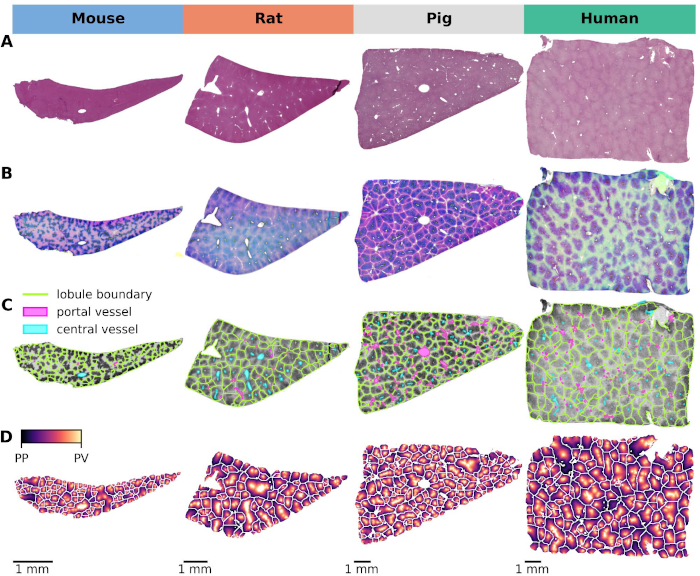
The mammalian liver is organized into three-dimensional structures called lobules that are integral to its function. According to the widely accepted hexagonal model, the cross-section of each lobule is characterized by a central vein and portal triads at the corners. These portal triads consist of a hepatic portal vein, a hepatic arteriole, and a bile duct. Blood is delivered through the hepatic artery and portal vein and flows inward from the outer periportal region toward the central vein through sinusoids lined with hepatocytes. These hepatocytes express a wide variety of metabolic enzymes, including cytochrome P450 (CYP) isoforms that are critical for xenobiotic metabolism. Notably, many hepatic enzymes, including CYPs, exhibit differential expression along the periportal-to-perivenous axis, a spatial variation known as liver zonation. Non-alcoholic fatty liver disease (NAFLD) is one of the most common liver diseases characterized by the accumulation of fat in the liver (steatosis) in the absence of significant alcohol consumption. Histopathologically, NAFLD is characterized by the presence of microvesicular fat droplets within hepatocytes. Although liver zonation has been extensively studied, the protein gradients of CYPs and their alterations in response to steatosis have not been systematically quantified or compared across species. This study aimed to develop an image analysis workflow to accurately derive these lobular expression gradients and quantify macrosteatosis in whole slide images (WSI) of liver histology. A comprehensive analysis was performed on whole slide images (WSIs) of mouse, rat, pig, and human liver specimens. These images included hematoxylin and eosin (H&E) stained slides and immunohistochemically stained slides for glutamine synthetase (GS) and cytochrome P450 isoforms 1A2, 2E1, 2D6, and 3A4. To achieve the study objectives, two image analysis workflows were developed: one using classical methods and the other using convolutional neural networks (CNN). These workflows were designed to perform two key tasks: (1) segmentation of liver lobules and (2) detection of macrosteatosis droplets. Key findings were that there was significant variability in lobular geometry within subjects, with strong consistency across species, with inter-species differences observed primarily in lobular size. A slight increase in lobular size was observed in steatotic samples, with a moderate to strong correlation between lobular size and steatosis content. In addition, interspecies differences in zonation patterns were observed for CYP1A2 in pigs, CYP2E1 in rats, and CYP3A4 in humans. Importantly, zonation patterns in steatotic subjects were not qualitatively different from those in non-steatotic subjects across species. In summary, I have successfully developed and implemented an image analysis workflow for whole slide images (WSIs) that allows detailed examination of liver lobule geometry and macrosteatosis droplets. Furthermore, the resulting datasets were integrated to facilitate a comprehensive analysis of the zonal distribution of CYP protein expression and macrosteatosis within the lobules.
2024-08-08
Open Science Ambassador of the BUA
Proud to be selected as Open Science Ambassador of the Berlin University Alliance (BUA) in 2024.
To find discipline-specific and accessible solutions to be implemented into everyday research routines, we have applied for Open Science Ambassador. As part of the work we will work on the reproducibility of computational models.
2024-07-18
Participation in DHPE
Matthias König selected for the Digital Health Professions Educator (DHPE) Program
We are excited to announce that Matthias König has been selected to participate in the "Digital Health Professions Educator" (DHPE) program for 2024/25. This program, part of the "HEDS: Handlungs- und Entscheidungskompetenz Digital Stärken" project, aims to enhance digital teaching skills at Charité. Running from June 2024 to June 2025, the program includes monthly in-person sessions and digital self-study units, leading to a nationally recognized certification (MQ II).
We are looking forward to embark on this journey to advance our expertise in digital health education!
2024-07-18
Humboldt Internship Program (HIP) conference
Proud to participate in Humboldt Internship Program 2024. Great work by Vera, Deepa and Prerna.
Students presented their results during the Humboldt Internship Program 2024. Great work by Vera, Deepa and Prerna.
2024-07-15
Final ecosystem meeting of EDITH on building the VHT
Proud to participate in the final ecosystem meeting of EDITH on building the virtual human twin (VHT) in Amsterdam.
The first day was dedicated to a plenary session on the EDITH proof of concept infrastructure, presented from the users’ perspective, where Researchers, Communities, together with Healthcare Providers and Patients were well represented to connect resources and enlighten the federated nature of the VHT, as well as to deploy VHT at bed side in clinical workflows and allow patients to upload their own data in the VHT, while representatives from Industry explored possible interaction of services within VHT.
2024-07-01
New project: Vera Tereshchuk starts SGLT2 PBPK Modeling Project
Vera Tereshchuk will develop a PBPK models for canagliflozin, aiming to enhance understanding of pharmacokinetics and pharmacodynamics in diabetes.
Vera Tereshchuk is developing a physiologically based pharmacokinetic/pharmacodynamic (PBPK/PD) model of the SGLT2 inhibitor canagliflozin. The objective is to enhance understanding of intraindividual variability in diabetes treatment with SGLT2 inhibitors, focusing on differences in hepatorenal function.
2024-06-25
PAGE2024: Presentation of our research in Rome
Mariia Myshkina and Matthias König presented our captopril modeling results during the PAGE2024 (Population Approach Group Europe) meeting in Rome.
Introduction: Angiotensin-converting enzyme (ACE) inhibitors, such as captopril, are widely prescribed for the treatment of hypertension and heart failure. Captopril, the first ACE inhibitor developed in the 1980s, is primarily taken orally and is rapidly absorbed, reaching peak plasma concentrations within an hour. Its half-life is approximately 2 hours, but several factors, including renal function, heart disease, age and sex, can affect its pharmacokinetics. Despite the widespread use of captopril, comprehensive pharmacokinetic data have been lacking. Our group has developed PK-DB (https://pk-db.com) [1], an open database containing high-quality pharmacokinetic data from clinical and preclinical research. PK-DB includes patient cohort characteristics, interventions, concentration-time profiles, and kinetic parameters, with the ability to automatically calculate pharmacokinetic parameters using non-compartmental methods. Objectives: The objectives of this study were to curate and analyze captopril pharmacokinetic data, expand the PK-DB, develop a whole-body physiologically based pharmacokinetic/pharmacodynamic (PBPK/PD) model of captopril in accordance with FAIR principles [2], and improve understanding of the intra-individual variability in captopril pharmacokinetics influenced by factors such as age, sex, renal and cardiac function. Methods: Over 600 publications on captopril pharmacokinetics were reviewed through a systematic literature search, with a subset selected for data curation in PK-DB. The PBPK model was developed using a compartmental approach and encoded in Systems Biology Markup Language (SBML) [3] for accessibility and reproducibility. Results: A comprehensive set of captopril pharmacokinetic data was generated from 15 clinical studies in healthy volunteers and patients with renal impairment, heart failure and hypertension. The PBPK model includes four major compartments (blood, intestine, liver, kidney) and describes absorption, distribution, metabolism and excretion. It successfully represents the pharmacokinetic profile of captopril in different age groups, showing minimal age-related differences in unchanged drug concentration. Heart failure did not significantly affect maximum plasma concentration or area under the curve (AUC), but renal function strongly influenced captopril elimination. We present this data set and open PBPK model of captopril as a valuable resource for further research. Conclusions: This study successfully establishes a freely accessible, comprehensive dataset and an open PBPK model of captopril, enhancing our understanding of its pharmacokinetic behavior across different patient groups. Our findings emphasize the significance of renal function in captopril elimination and provide a valuable resource for personalized medicine approaches in hypertension and heart failure treatment.
2024-06-09
Bachelor Thesis Afruja Hossain
Afruja Hossain finished her Bachelor thesis developing a database of CYP and UGT enzymes
Afruja Hossain has successfully finished her Bachelor thesis titled 'A Systematic Overview of Protein Variability in Cytochrome P450 and UDP-glucuronosyltransferase Enzymes in the Human Liver.' Her research provides an in-depth analysis of the key liver enzymes Cytochrome P450 (CYP450) and UDP-glucuronosyltransferase (UGT), crucial for drug metabolism. The study highlights the variability in protein levels of these enzymes, influenced by factors such as age, smoking, and alcohol consumption. Key findings include: - Creation of a database of CYP and UGT protein levels in the human liver. - Identification of significant individual variability in enzyme protein levels. - Correlations between different CYP and UGT isoforms. Afruja's work offers valuable insights into drug metabolism and could enhance understanding of drug interactions and toxicity. Congratulations to Afruja on her significant academic achievement.
2024-06-07
Successful Completion of One-Week Pharmacokinetic Modelling Course
We are excited to announce the successful completion of our one-week intensive course on Pharmacokinetic Modelling, where students delved into principles, drug distribution, clearance, elimination, and various modelling techniques to enhance drug therapy and development.
Pharmacokinetic modelling is the study of how drugs are absorbed, distributed, metabolised and excreted in the body. The pharmacokinetic modelling course covers topics such as pharmacokinetic principles, drug distribution, clearance and elimination and the factors that influence these processes. Students will learn about different models used to describe pharmacokinetics, such as compartmental models and physiologically based pharmacokinetic models, and how these models can be used to predict drug concentrations and optimise dosing regimens. Other topics that may be covered include pharmacodynamics, drug-drug interactions and the use of pharmacokinetic modelling in drug development and clinical practice. Overall, a course in pharmacokinetic modelling will provide students with a comprehensive understanding of the principles and techniques used to describe the movement of drugs through the body and how this knowledge can be applied to improve drug therapy.
2024-05-23
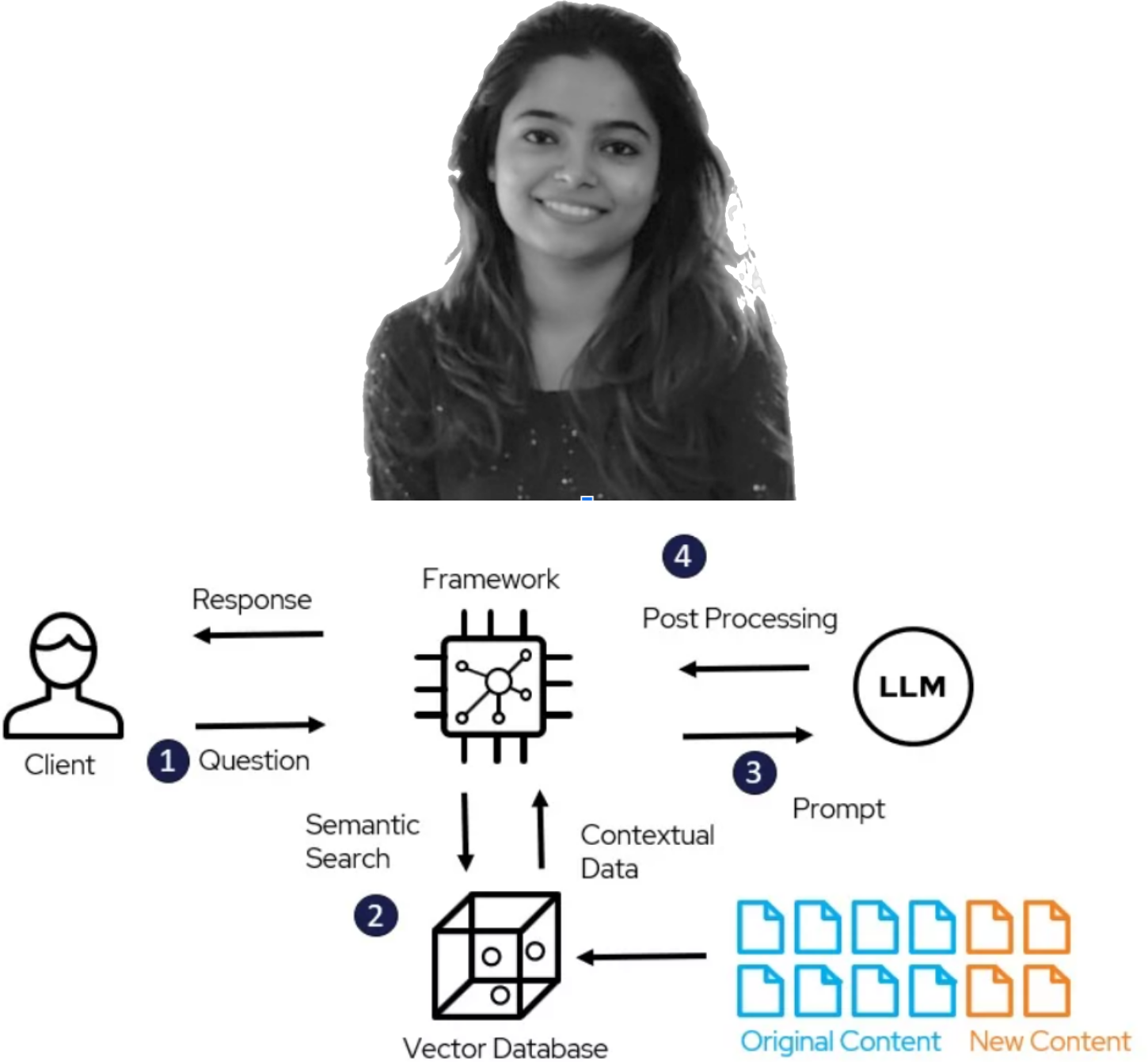
New Project Launch via Humboldt Internship Program (HIP): PK-LLM by Prerna Parakkat
We are excited to announce the start of a new project under the Humboldt Internship Program (HIP) by Prerna Parakkat. Prerna will be working on PK-LLM: Implementing a Large Language Model (LLM) as an Expert System for Pharmacokinetic (PK) Data Curation from Scientific Publications.
The project focuses on creating a specialized LLM tailored to interpret and extract pharmacokinetic data from diverse scientific texts, automating the extraction of key data such as parameters, dosing information, and subject details. It also aims to integrate this system with existing pharmacokinetic databases and computational modeling workflows for digital twins. We anticipate significant improvements in the accuracy and efficiency of pharmacokinetic data extraction and curation, setting a new standard in the field.
2024-05-16
Preprint: Cross-Species Variability in Lobular Geometry and Cytochrome P450 Hepatic Zonation: Insights into CYP1A2, CYP2E1, CYP2D6 and CYP3A4
Our latest publication on 'Cross-Species Variability in Lobular Geometry & Cytochrome P450 Hepatic Zonation' reveals insights into liver function across species using WSI technology.
This study explores the critical interplay between lobular geometry and the zonated distribution of cytochrome P450 (CYP) enzymes across species. We present an innovative approach to assess lobular geometry and zonation patterns using whole slide imaging (WSI). This method allows a detailed, systematic comparison of lobular structures and spatial distribution of key CYP450 enzymes and glutamine synthetase in four different species (mouse, rat, pig, and human). Our results shed light on species differences in lobular geometry and enzymatic zonation, providing critical insights for drug metabolism research. Based on our approach we could determine the minimum number of lobules required for a statistically representative analysis, an important piece of information when evaluating liver biopsies and deriving information from WSI.
2024-04-15
New projects: Nike Nemitz and Yusuf Ali Kulanoglu start PBPK Modeling Projects
Nike Nemitz and Yusuf Ali Kulanoglu will develop PBPK models for dapagliflozin and aliskiren, respectively, aiming to enhance understanding of pharmacokinetics and pharmacodynamics in diabetes and cardiovascular treatments.
Nike Nemitz is developing a physiologically based pharmacokinetic/pharmacodynamic (PBPK/PD) model of the SGLT2 inhibitor dapagliflozin. The objective is to enhance understanding of intraindividual variability in diabetes treatment with SGLT2 inhibitors, focusing on differences in hepatorenal function. Yusuf Ali Kulanoglu is working on a PBPK model of the direct renin inhibitor aliskiren to improve understanding of its pharmacokinetics and the influencing factors. Both projects aim to provide deeper insights into drug behavior and variability, potentially improving therapeutic outcomes.
2024-04-15
Publication: The Simulation Experiment Description Markup Language (SED-ML): Language Specification for Level 1 Version 5
Level 1 Version 5 of SED-ML enhances the ability to annotate, archive, share, and reproduce computational simulation experiments by allowing modelers to define tasks, model changes, ranges, and outputs using the Kinetic Simulation Algorithm Ontology (KiSAO).
We are excited to announce the latest publication titled "The Simulation Experiment Description Markup Language (SED-ML): Language Specification for Level 1 Version 5." This publication addresses the growing need in modern biological research for methods to annotate, archive, share, and reproduce computational simulation experiments. As computational simulations become more integral to biological research, extensive collaboration among modelers, experimentalists, and engineers is required. The Minimum Information About a Simulation Experiment (MIASE) guidelines provide a framework for sharing these experiments, and SED-ML offers a computer-readable format for the information outlined by MIASE. Level 1 Version 5 of SED-ML significantly enhances the capabilities for modelers by allowing the definition of tasks, model changes, ranges, and outputs using the Kinetic Simulation Algorithm Ontology (KiSAO). This builds on Version 4, which permitted defining simulations entirely with KiSAO. SED-ML growing ecosystem includes a wide range of investigators, model languages, and software tools, supporting various simulation tools, visual editors, model repositories, and validators. For more information about SED-ML and its applications, visit https://sed-ml.org.
2024-05-05
Master Thesis Shubhankar Palwankar
Shubhankar Palwankar defended his Master Thesis developing a PBPK model of enalapril
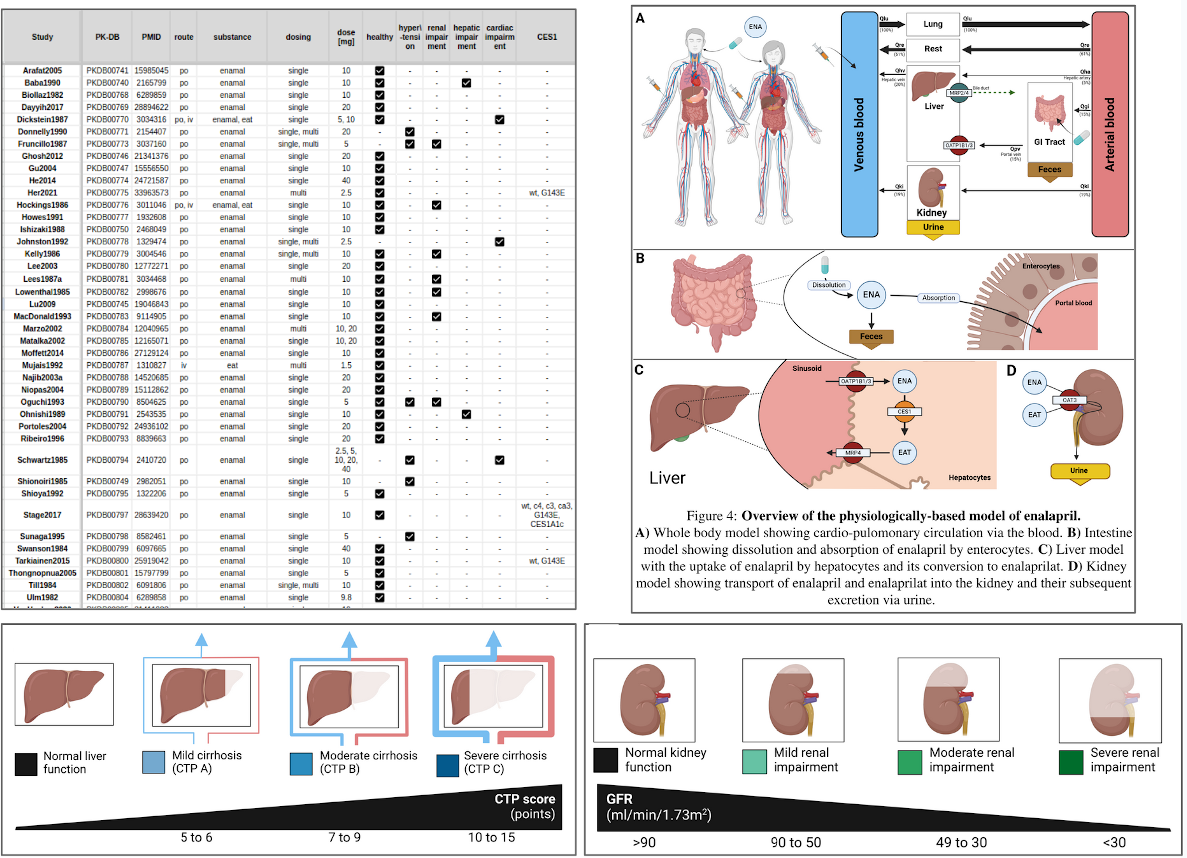
We are pleased to announce that Shubhankar Palwankar has successfully defended his Master Thesis, which focuses on the development of a Physiologically Based Pharmacokinetic (PBPK) model of enalapril, a key medication used to treat high blood pressure and other cardiovascular conditions. Enalapril works as an ACE inhibitor, playing a crucial role in regulating blood pressure through the renin-angiotensin-aldosterone system (RAAS). Shubhankar's research involved the creation of an extensive database from 49 clinical trials, which was used to parameterize and validate his computational model. His model effectively simulates the pharmacokinetics of enalapril and its active metabolite, enalaprilat, under various conditions including renal and hepatic impairment and changes in CES1 enzyme activity. The findings from this study offer valuable insights into how different physiological impairments affect the drug's behavior in the body, showing excellent agreement with clinical data. The developed model and data are publicly accessible in SBML format under a CC-BY 4.0 license from the PK-DB pharmacokinetics database, contributing to open science and further research in pharmacokinetics. Congratulations to Shubhankar Palwankar for his significant contributions to pharmacological research and his successful thesis defense.
2024-03-29
New multiscale liver model unveils zonation-function dynamics in Drug-Induced Liver Injury (DILI).
We're excited to announce the preprint of our latest research, 'Simulation of zonation-function relationships in the liver using coupled multiscale models: Application to drug-induced liver injury.' This study introduces a multiscale digital twin of the liver, integrating models from cellular to lobular scales, and sheds light on how liver zonation patterns influence drug-induced liver injury.
Multiscale modeling requires the coupling of models on different scales, often based on different mathematical approaches and developed by different research teams. This poses many challenges, such as defining interfaces for coupling, reproducible exchange of submodels, efficient simulation of the models, or reproducibility of results. Here, we present a multiscale digital twin of the liver that couples a partial differential equation (PDE)-based porous media approach for the hepatic lobule with cellular-scale ordinary differential equation (ODE)-based models. The models based on the theory of porous media describe transport, tissue mechanical properties, and deformations at the lobular scale, while the cellular models describe hepatic metabolism in terms of drug metabolism and damage in terms of necrosis. The resulting multiscale model of the liver was used to simulate perfusion-zonation-function relationships in the liver spanning scales from single cell to the lobulus. The model was applied to study the effects of (i) protein zonation patterns (metabolic zonation) and (ii) drug concentration dependence on spatially heterogeneous liver damage in the form of necrosis. Depending on the zonation pattern, different liver damage patterns could be reproduced, including periportal and pericentral necrosis as seen in drug-induced liver injury (DILI). Increasing the drug concentration led to an increase in the observed damage pattern. A key point for the success was the integration of domain-specific simulators based on standard exchange formats, i.e., libroadrunner for the high-performance simulation of ODE-based systems and FEBio for the simulation of the continuum-biomechanical part. This allows a standardized and reproducible exchange of cellular scale models in the Systems Biology Markup Language (SBML) between research groups.
2024-03-01
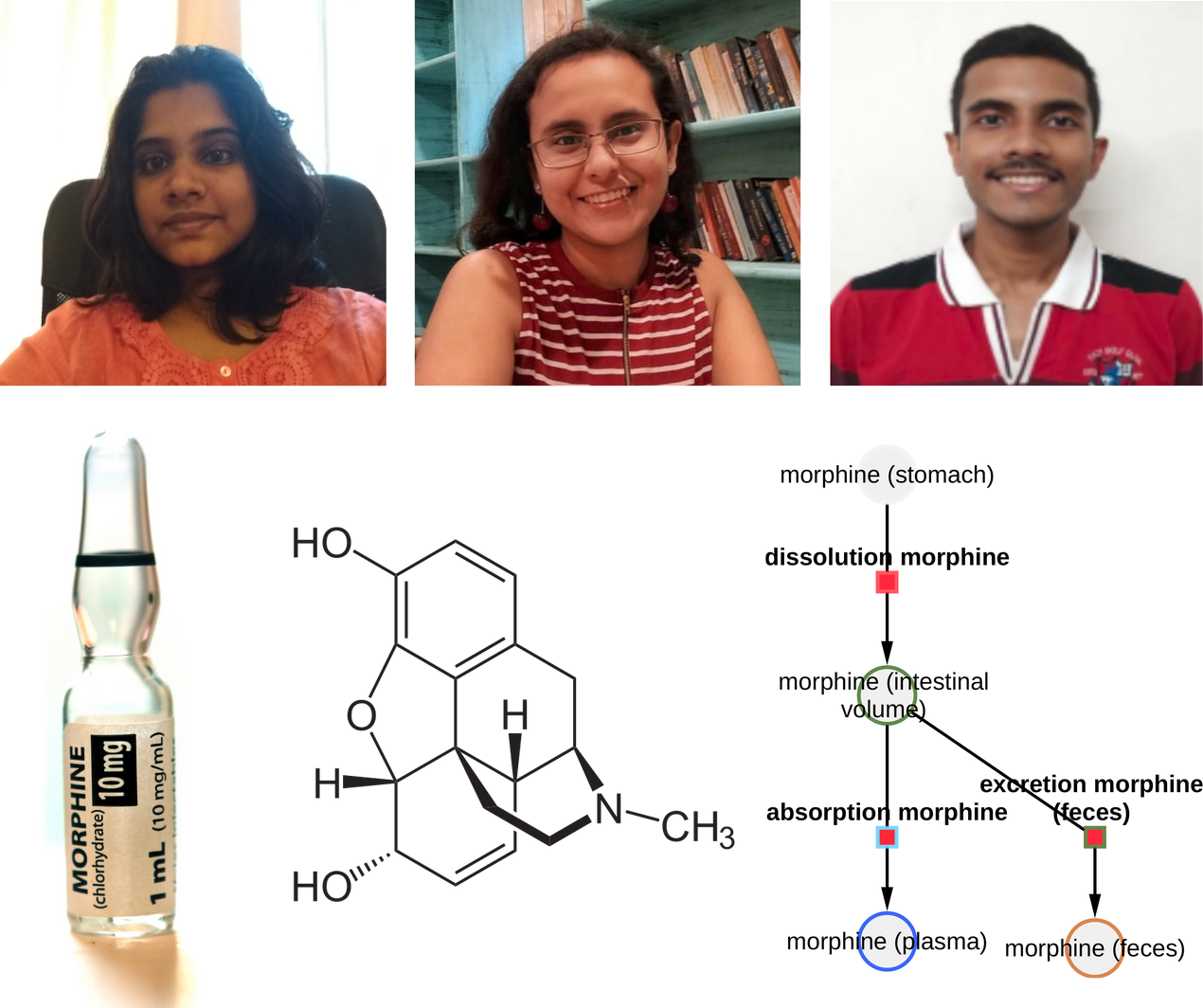
PBPK Model of Morphine Project
New Internship Project Launch: Development of a Physiologically Based Pharmacokinetics Model of Morphine. Deepa supported by Rohini and Rohit will develop a PBPK model of morphine.
Deepa supported by Rohini and Rohit will develop a physiologically based pharmacokinetic (PBPK) model of morphine. Morphine, a potent analgesic derived from the opium poppy, is essential for pain management but poses significant pharmacokinetic challenges. This project aims to address intra-individual variability in morphine treatment by establishing a comprehensive pharmacokinetics/pharmacodynamics database and constructing a whole-body computational model. Key areas of focus include the role of the liver and kidney in morphine metabolism and excretion, the effects of renal and hepatic impairment, and the effects of different morphine formulations and delivery methods. This research aims to improve the safe and effective use of morphine in clinical settings, taking into account factors such as age and genetic polymorphisms.
2024-02-01
Mariia Myshkina has started her PhD in the ATLAS project.
We welcome Mariia to our team who will be working on computational modeling of the liver within ATLAS.
Mariia will develop PBPK models of the liver in HCC coupled with machine learning approaches.
2024-01-18
Participation in the First EDITH-CSA Ecosystem Meeting - Advancing the Virtual Human Twin
We participated in the first EDITH-CSA 'Ecosystem meeting on building the Virtual Human Twin was held at the École Polytechnique de Paris on January 18 and 19, 2024.
After the official launch of the Virtual Human Twin initiative and the VHT Manifesto in December, the first open community meeting attracted over 180 registered guests from the EDITH-CSA consortium and the wider ecosystem. The enthusiasm of the wider community to travel to Paris (amidst difficult weather conditions) and proactively contributing towards developing the roadmap for the VHT was inspiring.
2023-12-29
Preprint: Cross-Species Variability in Lobular Geometry and Cytochrome P450 Hepatic Zonation: Insights into CYP1A2, CYP2E1, CYP2D6 and CYP3A4
Our latest preprint on 'Cross-Species Variability in Lobular Geometry & Cytochrome P450 Hepatic Zonation' reveals insights into liver function across species using WSI technology.
This study explores the critical interplay between lobular geometry and the zonated distribution of cytochrome P450 (CYP) enzymes across species. We present an innovative approach to assess lobular geometry and zonation patterns using whole slide imaging (WSI). This method allows a detailed, systematic comparison of lobular structures and spatial distribution of key CYP450 enzymes and glutamine synthetase in four different species (mouse, rat, pig, and human). Our results shed light on species differences in lobular geometry and enzymatic zonation, providing critical insights for drug metabolism research. Based on our approach we could determine the minimum number of lobules required for a statistically representative analysis, an important piece of information when evaluating liver biopsies and deriving information from WSI.
2023-11-03
Eleven Strategies for Making Reproducible Research and Open Science Training the Norm at Research Institutions
We are pleased to announce our latest paper in eLife that provides a roadmap for integrating reproducible research and open science practices into academic work.
We are excited to announce that our recent paper, "Eleven Strategies for Making Reproducible Research and Open Science Training the Norm at Research Institutions," has been published in eLife, offering a roadmap for integrating reproducible research and open science practices into academic work. Developed in collaboration with the German Reproducibility Network, the paper proposes eleven strategies focusing on modifying research assessment criteria, enhancing training, and fostering community support. This work aims to encourage researchers and institutions to adopt practices that increase the reliability and accessibility of scientific research.
2023-11-03
Your Dietary Digitial Twin (DDtwin)
Advancing Precision Nutrition: Workshop Success on Dietary Digital Twin, a workshop in Leiden from 30 October - 3 November 2023.
We are excited to announce our participation in a groundbreaking workshop dedicated to the development of the Dietary Digital Twin (DDtwin) technology platform. This innovative endeavor aims to provide precision nutrition by integrating biology-based and data-driven models with real-life data, harnessing the power of wearable sensors and applications.
The workshop brought together experts and enthusiasts from various fields to tackle the intricate task of extending models of whole-body glucose metabolism. The primary focus was on two technical challenges: the integration of real-time Continuous Glucose Monitoring (CGM) data from wearable sensors, and the creation of a human-centered user interface that connects the DDtwin to dietary intake data collected via apps.
The collaborative efforts made significant strides towards enabling the DDtwin to deliver personalized dietary insights and recommendations. Moreover, the workshop served as a platform for discussing the ethical, legal, and social implications (ELSI) of deploying such advanced technology in everyday practice.
Our participation in this workshop underscores our commitment to advancing health technology and contributing to a future where nutrition is as unique as the individual. Stay tuned for further developments as we continue to navigate the exciting realm of digital twins and precision nutrition. For more information see https://www.lorentzcenter.nl/your-dietary-digital-twin-ddtwin.html.
2023-10-31
Humboldt Internship Program 2024 - Computational Modeling of Drug Detoxification - A Systems Medicine Approach
We offer Internships in 2024, academic three-month research stays at Humboldt-Universität zu Berlin. The application for the Summer Term Internships 2024 is open from 03 November to 10 December 2023.
Humboldt Internship Program is an international short-term program for subject-specific, experiential learning. It allows participants to work with teams in research projects and university-related start-ups for three months.
This program is designed for advanced Bachelor as well as Master students and PhD candidates. Choose from projects in various academic areas, receive individual advising, and earn ECTS credit points. Some projects offer online internship opportunities. Please check the individual project descriptions for details.
The Summer Term internships take place from May to August 2023. More information https://hic.hu-berlin.de/en/internship-program .
2023-10-10
e:Med Meeting 2023 on Systems Medicine: Quantifying Fat Zonation in Liver Lobules: An Integrated Multiscale In-silico Model
We successfully presented our research on reproducible digital twins for personalized liver function assessment at the e:Med Meeting 2023 on Systems Medicine.
Essential prerequisites for the practical application and translation of computational models include: i) reproducibility of results; ii) model reusability and extensibility; iii) data availability; and iv) strategies for model stratification and individualization. Here, we present a modeling workflow built around these foundational prerequisites, with a focus on liver function tests.
Despite the paramount significance of liver function assessment in hepatology, reliable quantification remains a clinical challenge. Dynamic liver function tests offer a promising method for non-invasive in vivo assessment of liver function and metabolic phenotyping.
By leveraging whole-body physiologically-based pharmacokinetic (PBPK) models, we're simulating these tests and positioning PBPK models as digital twins for metabolic phenotyping and liver function assessment. To develop and validate our models, we established the open pharmacokinetics database, PK-DB, containing curated data from 600+ clinical studies. Our models are individualizable and stratifiable, enabling simulation of lifestyle factors and co-administration effects on drug metabolism. Our models have been instrumental in clinical scenarios: from predicting individual outcomes post-hepatectomy to discerning the impact of CYP2D6 gene variants on liver function tests. These models are constructed hierarchically, describing metabolic and other biological processes in organs like the liver and kidneys, seamlessly integrated with whole-body physiology.
Notably, all models and data are readily available and reproducible for reuse, encoded in the Systems Biology Markup Language (SBML). We will provide an overview of these PBPK models and demonstrate how SBML and FAIR principles can facilitate model development, coupling, and reuse.
2023-09-18
Preprint: Quantifying Fat Zonation in Liver Lobules: An Integrated Multiscale In-silico Model
Our latest preprint is now out: An integrated multiscale model for quantifying fat zonation in liver lobules.
We have developed a sophisticated computer model to explore how various factors contribute to the distribution and accumulation of fat in the liver, a phenomenon known as "metabolic zonation." This model will help in understanding how liver health is affected by different conditions and could play a crucial role in studying diseases like MASLD (metabolic dysfunction-associated steatotic liver disease). By simulating liver functions, the model considers interactions between blood flow, oxygen levels, and fat metabolism within liver lobules.
2023-08-28
Hannah Menghis from Brown University starts her internship
We welcome Hannah Menghis who will establish a PBPK model of semaglutide.
Hannah will establish a pharmacokinetic dataset of semaglutide and develop an initial version of a physiologically based pharmacokinetic (PBPK) model of the GLP-1 inhibitor semaglutide. The objective is to enhance our understanding of the underlying causes of intraindividual variability in semaglutide treatment, e.g., differences in renal function.
2023-08-02
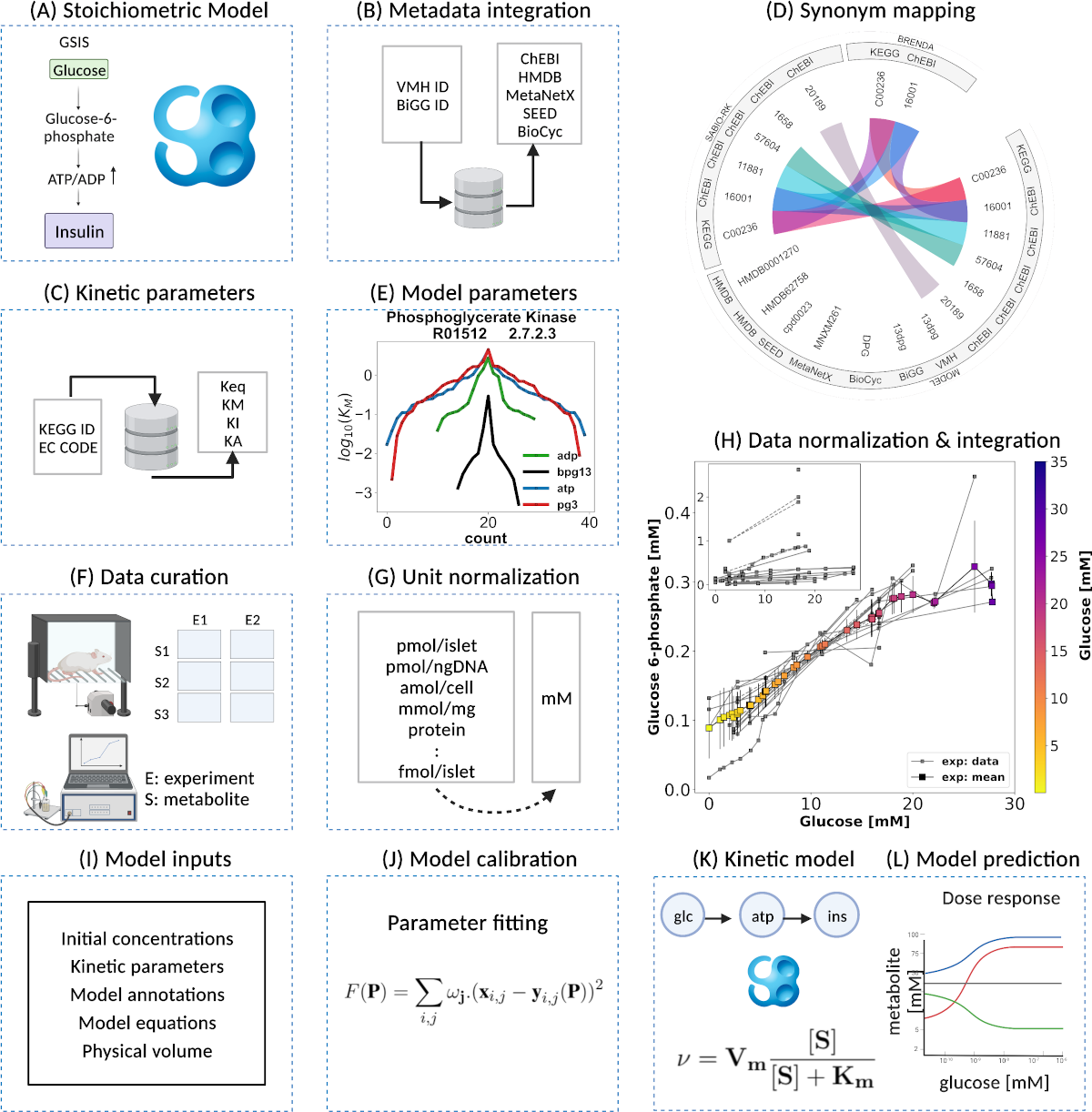
A pathway model of glucose-stimulated insulin secretion in the pancreatic β-cell
Our latest publication is now on Frontiers of Endocrinology: A pathway model of glucose-stimulated insulin secretion in the pancreatic β-cell
The pancreas plays a critical role in maintaining glucose homeostasis through the secretion of hormones from the islets of Langerhans. Glucose-stimulated insulin secretion (GSIS) by the pancreatic β-cell is the main mechanism for reducing elevated plasma glucose. Here we present a systematic modeling workflow for the development of kinetic pathway models using the Systems Biology Markup Language (SBML). Steps include retrieval of information from databases, curation of experimental and clinical data for model calibration and validation, integration of heterogeneous data including absolute and relative measurements, unit normalization, data normalization, and model annotation. An important factor was the reproducibility and exchangeability of the model, which allowed the use of various existing tools. The workflow was applied to construct a novel data-driven kinetic model of GSIS in the pancreatic β-cell based on experimental and clinical data from 39 studies spanning 50 years of pancreatic, islet, and β-cell research in humans, rats, mice, and cell lines. The model consists of detailed glycolysis and phenomenological equations for insulin secretion coupled to cellular energy state, ATP dynamics and (ATP/ADP ratio). Key findings of our work are that in GSIS there is a glucose-dependent increase in almost all intermediates of glycolysis. This increase in glycolytic metabolites is accompanied by an increase in energy metabolites, especially ATP and NADH. One of the few decreasing metabolites is ADP, which, in combination with the increase in ATP, results in a large increase in ATP/ADP ratios in the β-cell with increasing glucose. Insulin secretion is dependent on ATP/ADP, resulting in glucose-stimulated insulin secretion. The observed glucose-dependent increase in glycolytic intermediates and the resulting change in ATP/ADP ratios and insulin secretion is a robust phenomenon observed across data sets, experimental systems and species. Model predictions of the glucose-dependent response of glycolytic intermediates and biphasic insulin secretion are in good agreement with experimental measurements. Our model predicts that factors affecting ATP consumption, ATP formation, hexokinase, phosphofructokinase, and ATP/ADP-dependent insulin secretion have a major effect on GSIS. In conclusion, we have developed and applied a systematic modeling workflow for pathway models that allowed us to gain insight into key mechanisms in GSIS in the pancreatic β-cell.
2023-07-17
Bachelor Thesis Beatrice Stemmer Mallol
Beatrice Stemmer Mallol submitted her Bachelor thesis developing a PBPK model of talinolol
Congratulations to Bea for successfully submitting her Bachelor thesis on a physiologically based pharmacokinetic (PBPK) model of the probe drug talinolol for the characterization of intestinal P-glycoprotein. Talinolol, a cardioselective beta-blocker applied in treating various cardiovascular diseases and tachyarrhythmias, is widely adopted as a probe drug for the intestinal efflux transporter P-glycoprotein. In the human body, P-gp plays a crucial role, given its distribution across various tissues to protect against potentially toxic substances, while expediting the elimination of xenobiotics. By employing talinolol for P-gp phenotyping, researchers can evaluate factors that influence P-gp-mediated transport, such as P-gp genetic polymorphisms and its distribution throughout the intestine. Bea's thesis presents a comprehensive dataset on talinolol pharmacokinetics, the study of how the body absorbs, distributes, metabolizes, and excretes drugs. This data was used to develop a PBPK model for talinolol, offering detailed insights into its behaviour within the body. The new model investigates the impact of numerous factors on talinolol's pharmacokinetics, including the genetic variants of P-gp, enzymatic activity of transporters OATP2B1 and OATP1B1, and site-specific distribution of P-gp and OATP2B1 proteins in the intestine. It also investigates the impact of diseases like liver cirrhosis and renal dysfunction.
2023-07-07
PhD Defense of Jan Grzegorzewski
Jan Grzegorzewski defended his PhD thesis on physiologically based pharmacokinetic modeling (PBPK) for dynamical liver function tests and Cytochrome P450 (CYP) phenotyping.
Congratulations to Jan Grzegorzewski for successfully defending his PhD thesis on physiologically based pharmacokinetic modeling (PBPK) for dynamical liver function tests and Cytochrome P450 (CYP) phenotyping. Jan developed the extensive open pharmacokinetics database, PK-DB, which helped to demonstrate demographic biases and errors in existing pharmacokinetic literature. His research includes a meta-analysis on caffeine pharmacokinetics, discovering multiple factors influencing liver function and CYP1A2 activity. Furthermore, he developed a PBPK model to predict individual plasma concentrations of dextromethorphan based on the CYP2D6 genotype and physiological traits. His work is expected to significantly enhance the precision of dynamic liver function tests and advance personalized medicine.
2023-07-01
Xenia Petukhova and Zahra Bahri start Humboldt Internship Program
Humboldt Internship Program 2023 - Computational Modeling of Drug Detoxification - A Systems Medicine Approach
We welcome Xenia and Zhara in our group for the Humboldt Internship Program 2023. Xenia and Zhara will develop a physiologically based pharmacokinetic (PBPK) model for inulin and creatinine to assess the glomerular filtration rate (GFR). Our goal is to gain a deeper understanding of the impact of various inulin protocols on GFR measurement and the influence of physiological factors, such as body composition, on GFR determination using inulin. Furthermore, it should enhance our understanding of how variations in creatinine production, muscle mass, and tubular secretion of creatinine influence eGFR calculations.
2023-06-16
Keynote: Advancing Liver Function Assessment: Personalized and Stratified Approaches with Standardized Computational Models and Data
Keynote talk at the Workshop on Computational Models in Biology and Medicine 2023 organized by Nicole Radde and Sebastian Höpfl.
Essential prerequisites for the practical application and translation of computational models include: i) reproducibility of results; ii) reusability and extensibility of models; iii) data availability; and iv) strategies for model stratification and individualization. In this study, we present a modeling workflow tailored to these critical aspects, with a focus on liver function tests. Evaluating liver function is a crucial task in hepatology, yet accurately quantifying hepatic function has persisted as a clinical challenge. Dynamic liver function tests offer a promising method for non-invasive in vivo assessment of liver function and metabolic phenotyping. These clinical tests determine liver function through the elimination of a specific test substance, thus revealing information about the liver's metabolic capacity. We employed whole-body physiologically-based pharmacokinetic (PBPK) models to simulate these tests, which encompass absorption, distribution, metabolism, and elimination processes. PBPK models serve as powerful instruments for investigating drug metabolism and its impact on the human body. In this research, we showcase our efforts in utilizing PBPK models as digital twins for metabolic phenotyping and liver function evaluation. To develop and validate our models, we created the first open pharmacokinetics database, PK-DB, containing curated data from over 600 clinical studies. Our models are individualizable and stratifiable, enabling simulation of lifestyle factors and co-administration effects on drug metabolism. We have applied our models to various clinical inquiries, such as simulating individual outcomes post-hepatectomy using an indocyanine green model and examining the influence of CYP2D6 gene variants through a dextromethorphan model integrated with drug-gene interactions. These models are constructed hierarchically, describing metabolic and other biological processes in organs like the liver and kidneys, connected to whole-body physiology. All models and data are accessible for reuse in a reproducible manner, encoded in the Systems Biology Markup Language (SBML). In this study, we provide an overview of PBPK models and demonstrate how SBML, COMBINE standards, and FAIR principles can facilitate model development, coupling, and reuse.
2023-05-28
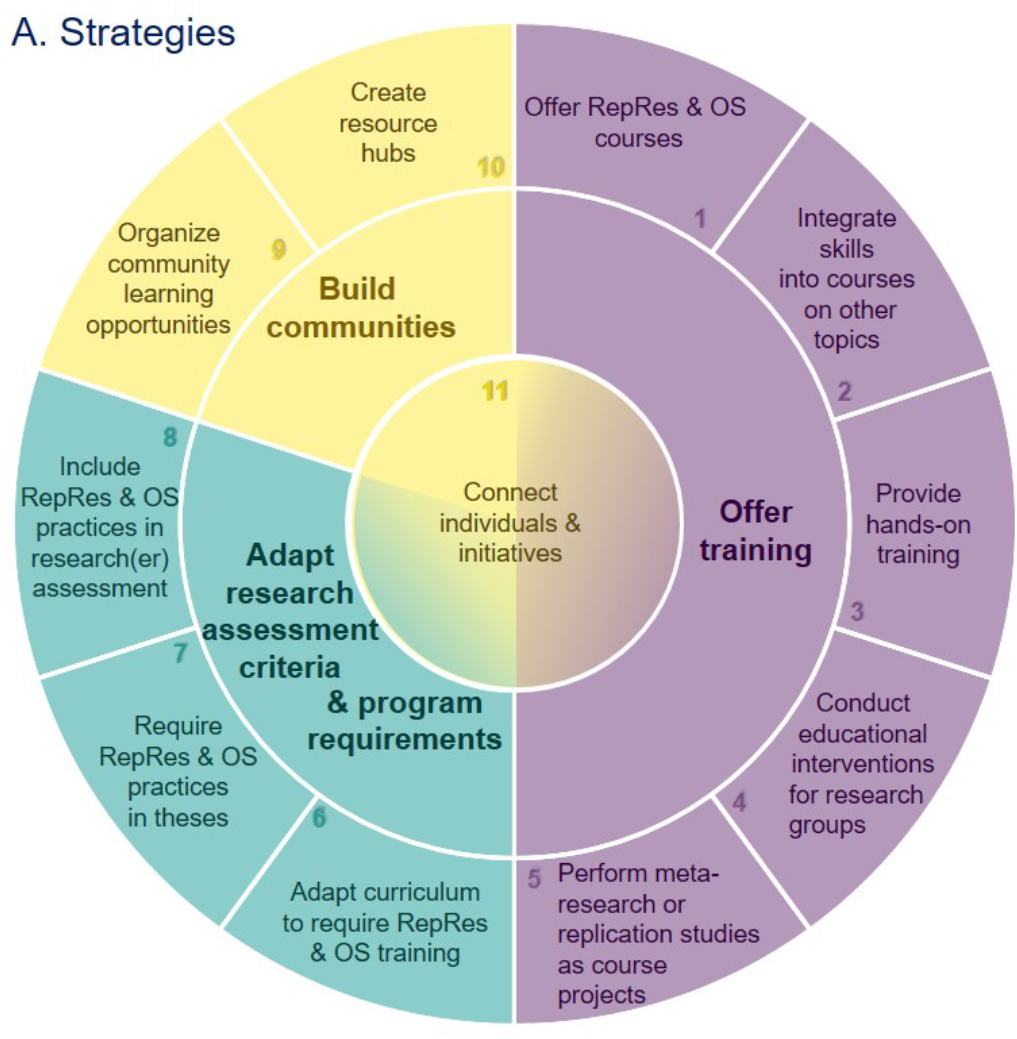
Preprint: Eleven Strategies for Making Reproducible Research and Open Science Training the Norm at Research Institutions
We present 11 strategies for making #OpenScience & #ReproducibleResearch the norm at research institutions, with tips for implementation & resources. Let's join hands to make these practices standard in the scientific community!
Across disciplines, researchers increasingly recognize that open science and reproducible research practices may accelerate scientific progress by allowing others to reuse research outputs and by promoting rigorous research that is more likely to yield trustworthy results. While initiatives, training programs, and funder policies encourage researchers to adopt reproducible research and open science practices, these practices are uncommon in many fields. Researchers need training to integrate these practices into their daily work. We organized a virtual brainstorming event, in collaboration with the German Reproducibility Network, to discuss strategies for making reproducible research and open science training the norm at research institutions. Here, we outline eleven strategies, concentrated in three areas: (1) offering training, (2) adapting research assessment criteria and program requirements, and (3) building communities. We provide a brief overview of each strategy, offer tips for implementation, and provide links to resources. Our goal is to encourage members of the research community to think creatively about the many ways they can contribute and collaborate to build communities, and make reproducible research and open science training the norm. Researchers may act in their roles as scientists, supervisors, mentors, instructors, and members of curriculum, hiring or evaluation committees. Institutional leadership and research administration and support staff can accelerate progress by implementing change across their institutions.
2023-04-16
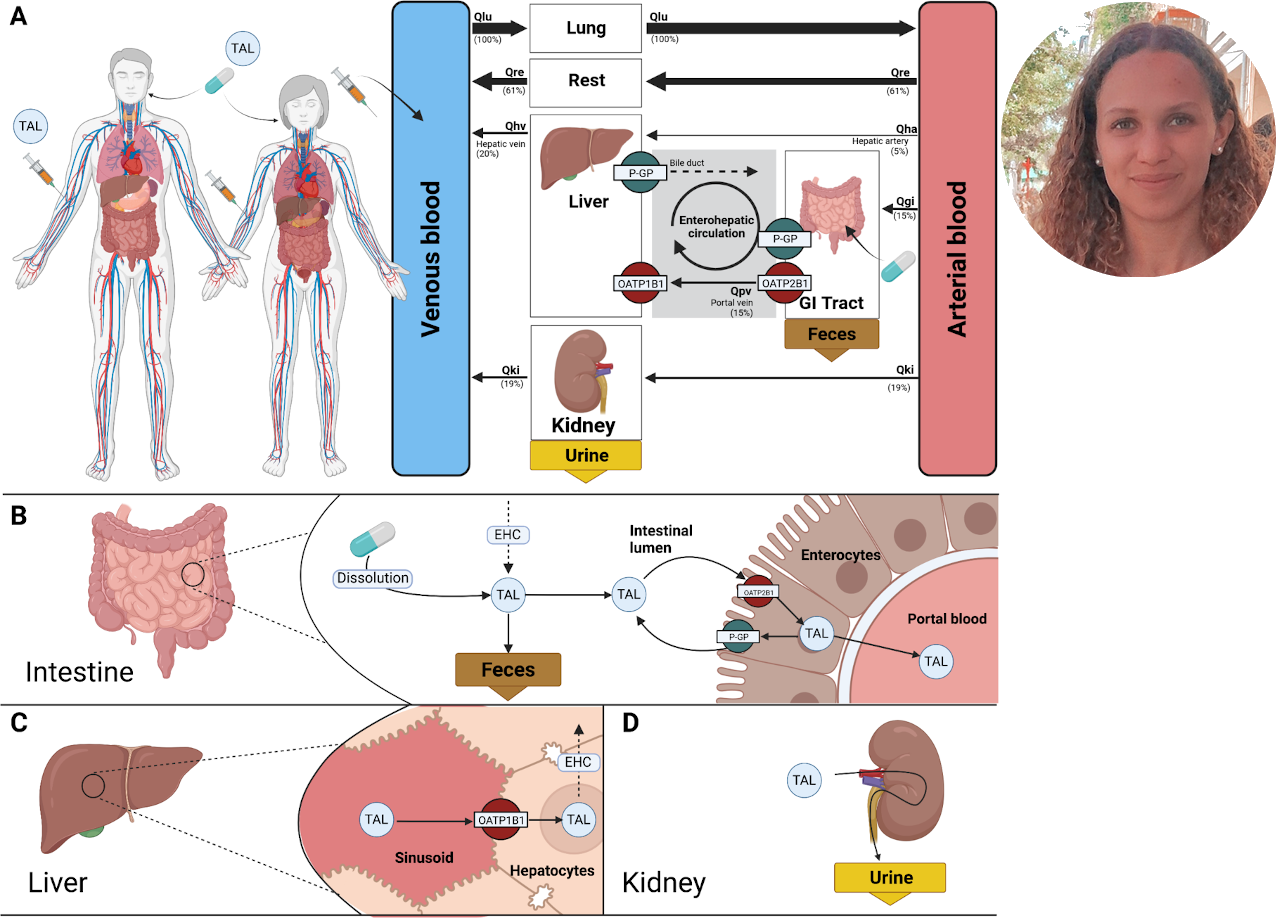
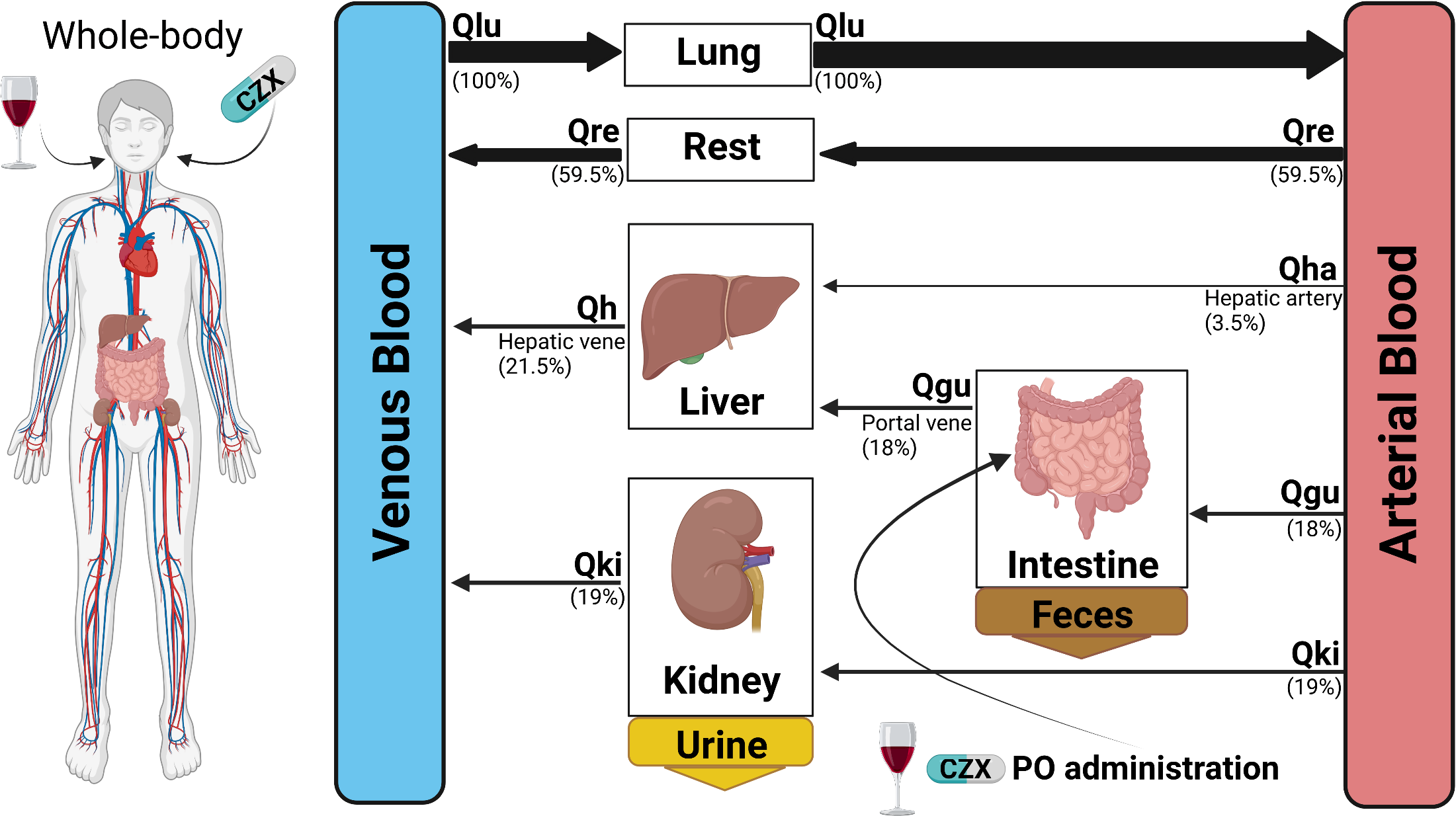
Preprint: A physiologically based pharmacokinetic model for CYP2E1 phenotyping via chlorzoxazone
Our latest preprint is now on bioRxiv: A physiologically based pharmacokinetic model for CYP2E1 phenotyping via chlorzoxazone
We have developed a physiologically based pharmacokinetic (PBPK) model for CYP2E1 phenotyping via chlorzoxazone. This model takes into account various factors that can influence the activity of CYP enzymes, such as genetics, diet, age, environmental factors, and disease, making it a highly effective and reliable tool for assessing the in vivo activity of the CYP2E1 enzyme. What is truly exciting about this research is that it incorporates the effect of ethanol consumption and liver impairment on the results of metabolic phenotyping with chlorzoxazone. This means that the model is not only accurate but also highly practical, as it takes into account factors that are commonly encountered in clinical practice. The extensive pharmacokinetic dataset for chlorzoxazone and the validated PBPK model are expected to have a significant impact on the field of pharmacokinetics, as they provide a reliable and comprehensive tool for assessing the activity of CYP2E1 in a wide range of clinical contexts. This research represents a major step forward in our understanding of the factors that influence the activity of CYP enzymes and promises to have a significant impact on the development of new drugs and the optimization of existing ones.
2023-04-03
Michael Stifel Price to promote interdisciplinary, data-driven research
M. Albadry, S. Höpfl, and M. König won the QuaLiPerF Michael Stifel Price for interdisciplinary, data-driven research.
M. Albadry, S. Höpfl, and M. König won the QuaLiPerF Michael Stifel Price for interdisciplinary, data-driven research for the study
Albadry M, Höpfl S, Ehteshamzad N, König M, Böttcher M, Neumann J, Lupp A, Dirsch O, Radde N, Christ B, Christ M, Schwen LO, Laue H, Klopfleisch R, Dahmen U. Periportal steatosis in mice affects distinct parameters of pericentral drug metabolism. Sci Rep. 2022 Dec 17;12(1):21825. doi: 10.1038/s41598-022-26483-6.
The study was conducted as part of a project to model the pharmacokinetics of steatotic mice and explore the impact of periportal steatosis on drug metabolism parameters. With the interdisciplinary approach of QuaLiPerF, we collaborated with three fields within life sciences: Mohamed Albadry for data generation, Matthias König for data analysis, and Sebastian Höpfl for uncertainty quantification. The data generation involved performing animal experiments, pharmacokinetic studies, and analyzing steatosis by quantifying its spatial distribution, zonated expression, and activity of drug metabolism enzymes (CYP).
2023-03-29
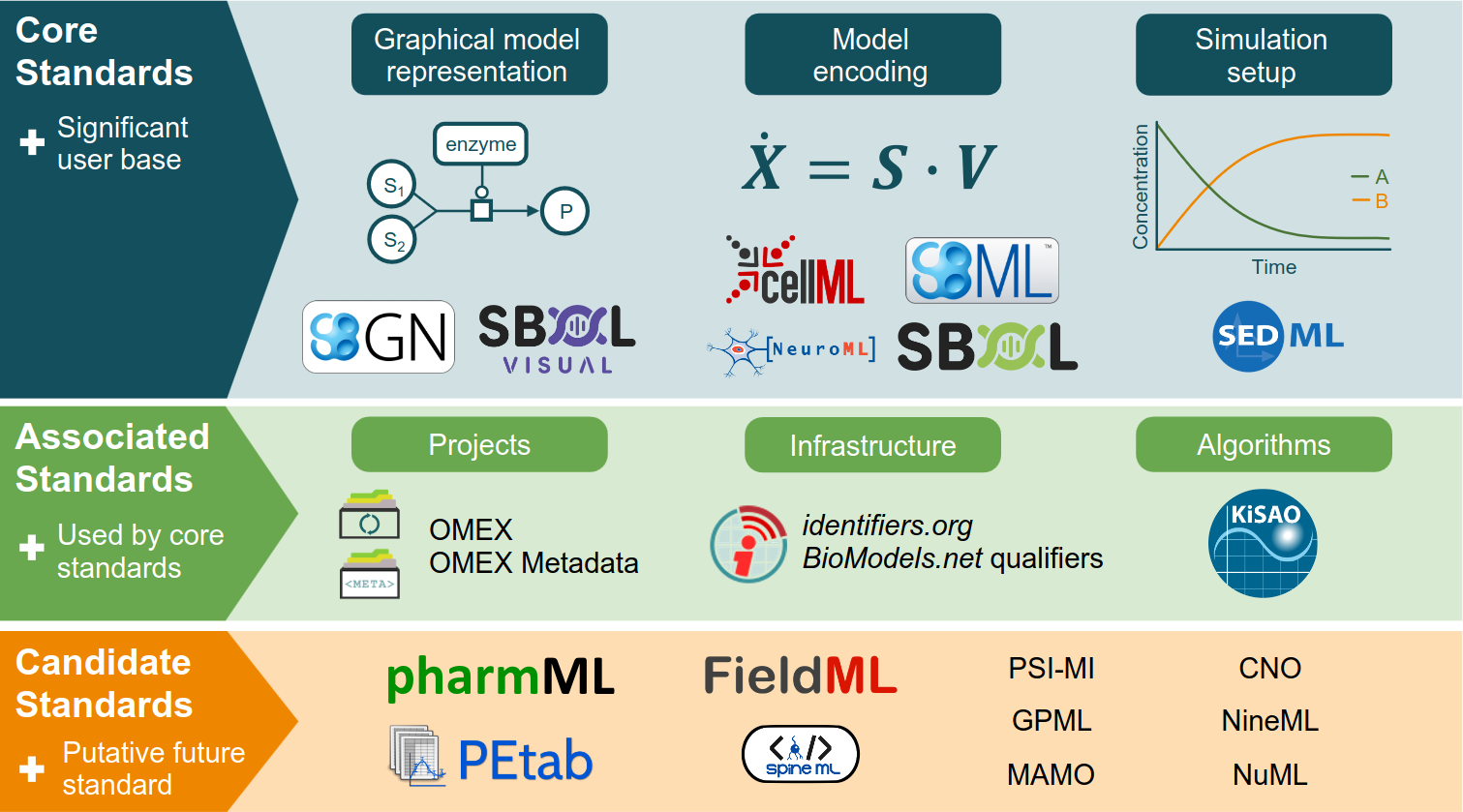
Publication: Specifications of standards in systems and synthetic biology: status and developments in 2022 and the COMBINE meeting 2022
Our overview of specifications in systems and synthetic biology with a brief overview of the COMBINE 2022 is available.
The Journal of Integrative Bioinformatics has published our editorial for a special issue providing updated specifications of the COMBINE standards in systems and synthetic biology. The 2022 issue includes three updates to the standards, namely CellML 2.0.1, SBML Level 3 Package: Spatial Processes, Version 1, Release 1, and Synthetic Biology Open Language (SBOL) Version 3.1.0. This publication is a valuable resource for researchers looking to keep up with the latest developments in COMBINE standards. Additionally, the editorial section of the issue provides a brief overview of the COMBINE 2022 meeting held in Berlin.
2023-03-13
Preprint: A consensus model of glucose-stimulated insulin secretion in the pancreatic beta-cell
Our latest preprint is now on bioRxiv: A consensus model of glucose-stimulated insulin secretion in the pancreatic beta-cell
The pancreas plays a critical role in maintaining glucose homeostasis through the secretion of hormones from the islets of Langerhans. Glucose-stimulated insulin secretion (GSIS) by the pancreatic beta-cell is the main mechanism for reducing elevated plasma glucose. Here we present a systematic modeling workflow for the development of kinetic pathway models using the Systems Biology Markup Language (SBML). The workflow was applied to construct the first consensus model of GSIS in the pancreatic beta-cell based on experimental and clinical data from 39 studies spanning 50 years of pancreatic, islet, and beta-cell research in humans, rats, mice, and cell lines. The model consists of detailed glycolysis and equations for insulin secretion coupled to cellular energy state (ATP/ADP ratio). Key findings of our work are that in GSIS there is a glucose-dependent increase in almost all intermediates of glycolysis. This increase in glycolytic metabolites is accompanied by an increase in energy metabolites, especially ATP and NADH. One of the few decreasing metabolites is ADP, which, in combination with the increase in ATP, results in a large increase in ATP/ADP ratios in the beta-cell with increasing glucose. Insulin secretion is dependent on ATP/ADP, resulting in glucose-stimulated insulin secretion. The observed glucose-dependent increase in glycolytic intermediates and the resulting change in ATP/ADP ratios and insulin secretion is a robust phenomenon observed across data sets, experimental systems and species.
2023-02-12
PBPK modelling of sorafenib and Gd-EOB-DTPA
Frances Okibedi and Narudeem Sojimade will develop models of the anticancer drug sorafenib in HCC and the liver contrast agent Gd-EOB-DTPA.
Within this project, a high quality database of sorafenib pharmacokinetic data used in systemic therapy of hepatocellular carcinoma will be established. Based on the pharmacokinetic data set, a physiologically based pharmacokinetic (PBPK) model will be established for the evaluation of systemic sorafenib therapy in liver cancer. The model will be used to address important clinically relevant questions such as (1) sorafenib is contraindicated in advanced liver cirrhosis, (2) genetic variants of OATP uptake transporters may alter the efficacy of sorafenib, (3) CYP3A4 substrates lead to drug-drug interactions with sorafenib.
Using established data curation pipelines, this project will extend our pharmacokinetics database PK-DB with literature data on Gd-EOB-DTPA. Based on the pharmacokinetic data set, a physiologically based pharmacokinetic (PBPK) model will be established for the evaluation of the pharmacokinetics of Gd-EOB-DTPA and its use as an MRI contrast agent. The model will be used to address important clinically relevant questions such as (1) the effect of renal and hepatic impairment on Gd-EOB-DTPA pharmacokinetics, (2) the effect of perfusion and OATP on Gd-EOB-DTPA pharmacokinetics, (3) the relationship between concentration and MRI attenuation.
2023-02-12
X-Research Group - Digital twins for the treatment of hypertension
M. König receives funding of the Berlin University Alliance and the DFG for the X-Student Research Group: Digital twins for the treatment of hypertension
Understanding a drugs pharmacokinetics - how it is absorbed, distributed, metabolised and excreted by the body - and its pharmacodynamics - how it affects the body - is a key challenge in treating people. Angiotensin converting enzyme (ACE) inhibitors and diuretics are two classes of drugs used to treat high blood pressure. Both are among the most commonly prescribed drugs due to the high prevalence of hypertension in an ageing society.
In this X-student research group we will develop physiologically based pharmacokinetic models of the diuretic hydrochlorothiazide and the ACE inhibitor lisinopril. The methodological approach is a combination of lectures, tutorials and practical work by the students. The research project is aimed at students with a STEM (science, technology, engineering, mathematics) background, including biology, computer science and medical students. Basic programming skills in Python are required.
With the X-Student Research Groups, the Berlin University Alliance supports research teams consisting of young researchers and students. The goal is to involve students in current research projects of the alliance partners and to enable them to participate in (cutting-edge) research already during their studies.
Funded under the Excellence Strategy of the Federal Government and the Länder by the Berlin University Alliance.
2023-02-07
M. König has been elected as PEtab editor (2023-2026)
Proud to be elected editor for PEtab, a data format for specifying parameter estimation problems in systems biology.
PEtab is a data format for specifying parameter estimation problems in systems biology. PEtab is built around SBML and is based on tab-separated values (TSV) files. It is intended as a standardized way to provide information for parameter estimation that is beyond the current scope of SBML. This includes, for example: (1) specifying and linking measurements to models; (2) defining model outputs; (3) specifying noise models; (4) specifying parameter bounds for optimization; (5) specifying multiple simulation conditions with potentially shared parameters. Matthias König has been elected PEtab Editor from March 2023 to March 2026.
2023-02-03
Preprint: Simvastatin therapy in different subtypes of hypercholesterolemia
Our latest preprint is now on medRxiv: Simvastatin therapy in different subtypes of hypercholesterolemia - a physiologically based modelling approach.
Hypercholesterolemia is a multifaceted plasma lipid disorder with heterogeneous causes including lifestyle and genetic factors. A key feature of hypercholesterolemia is elevated plasma levels of low-density lipoprotein cholesterol (LDL-C). Several genetic variants have been reported to be associated with hypercholesterolemia, known as familial hypercholesterolemia (FH). Important variants affect the LDL receptor (LDLR), which mediates the uptake of LDL-C from the plasma, apoliporotein B (APOB), which is involved in the binding of LDL-C to the LDLR, and proprotein convertase subtilisin/kexin type 9 (PCSK9), which modulates the degradation of the LDLR. A typical treatment for hypercholesterolemia is statin medication, with simvastatin being one of the most commonly prescribed statins. In this work, the LDL-C lowering therapy with simvastatin in hypercholesterolemia was investigated using a computational modeling approach. A physiologically based pharmacokinetic model of simvastatin integrated with a pharmacodynamic model of plasma LDL-C (PBPK/PD) was developed based on extensive data curation. A key component of the model is LDL-C turnover by the liver, consisting of: hepatic cholesterol synthesis with the key enzymes HMG-CoA reductase and HMG-CoA synthase; cholesterol export from the liver as VLDL-C; de novo synthesis of LDLR; transport of LDLR to the membrane; binding of LDL-C by LDLR via APOB; endocytosis of the LDLR-LDL-C complex; recycling of LDLR from the complex. The model was applied to study the effects of simvastatin therapy in hypercholesterolemia due to different causes in the LDLR pathway corresponding to different subtypes of hypercholesterolemia. Model predictions of LDL-C lowering therapy were validated with independent clinical data sets. Key findings are: (i) hepatic LDLR turnover is highly heterogeneous among FH classes; (ii) despite this heterogeneity, simvastatin therapy results in a consistent reduction in plasma LDL-C regardless of class; and (iii) simvastatin therapy shows a dose-dependent reduction in LDL-C. Our model suggests that the underlying cause of hypercholesterolemia does not influence simvastatin therapy. Furthermore, our model supports the treatment strategy of stepwise dose adjustment to achieve target LDL-C levels. Both the model and the database are freely available for reuse.
2023-02-01
ATLAS - AI and Simulation for Tumor Liver ASsessment
The BMBF is funding our joint project ATLAS in the "Computational Life Sciences - AI Methods for Systems Medicine".
Liver cancer is the second most common cause of cancer-related death. Diagnosis and treatment are time-critical and require highly patient-specific diagnostic and treatment pathways. Medical decision-making is based on a variety of interdependent factors related to different medical disciplines, past experience and clinical guidelines. Taking into account all decision factors in combination with the possible therapeutic approaches is a major challenge for physicians and often cannot be solved optimally even in an interdisciplinary tumor board. In this project, we are developing ATLAS, a decision support tool that will significantly assist clinicians in meeting this challenge. Based on AI methods, ATLAS processes all relevant patient data from databases, systems medicine and continuum biomechanical in silico prognosis models as well as individual patient data. The tool is being developed in a co-design approach by experts in surgical oncology, mathematical modeling and machine learning. The selected technologies will integrate automated understanding of a highly complex patient situation through simulation of liver functions with expert knowledge and ontology-driven learning with knowledge graphs from retrospective liver tumor cases. ATLAS will be based on a detailed historical data cohort of more than 6,000 patients with liver tumors and will be evaluated on case studies at the University Hospital of Jena. The integration of medical expert knowledge, mathematical modeling and artificial intelligence represents a highly original and promising approach for high-quality diagnosis and treatment of liver tumors, resulting in patient-specific improvement of prognosis. The scientific knowledge gained from these projects will provide opportunities for transfer to malignancies in other organs, such as the lung, kidney or brain. The development of tools and demonstrators will provide sustainable exploitation pathways for future commercial applications.
2022-11-17
New Bachelor Student - Afruja Hossain
Afruja Hossain started working in our lab on a systematic overview of the protein variability of cytochrome P450 (CYP450) isoforms in the human liver.
2022-11-15
Visiting EU parliament and attending STOA AI session
We were selected to participate in an European policy journey on the topic EU research funding and science communication by the Berlin University Alliance (BUA).
2022-10-26
Humboldt Internship Program 2023 - Computational Modeling of Drug Detoxification - A Systems Medicine Approach
https://hic.hu-berlin.de/en/internship-program/projects/2012022-10-24
Dextromethorphan publication
How does CYP2D6 genotype effect metabolic phenotyping? See our latest paper in Frontiers in Pharmacology for answers.
2022-10-24
X-Research Group
M. König receives funding of the Berlin University Alliance and the DFG for the X-Student Research Group: Physiologically based modeling of drugs: ACE inhibitors
2022-10-06
COMBINE 2022
We organized the Computational Modeling in Biology Network (COMBINE) meeting in Berlin this year with > 100 participants from 06-08 October 2022.
2022-10-10
ICSB 2022 - Presentation and Posters
We could present our work on physiologically based pharmacokinetics (PBPK) modeling at ICSB2022.