
News
2025-04-01
SBML editor

|
Matthias König elected as an SBML Editor for the 2025–2027 term. |
2025-04-02
Featured Fellow - Berlin University Alliance (BUA) - Matthias König

|
As a featured BUA Fellow at Humboldt-Universität, Matthias König advances digital medicine and systems biology at the intersection of data, modeling, and clinical research. |
2025-02-21
Preprint: From FAIR to CURE: Guidelines for Computational Models of Biological Systems
|
Our latest preprint, From FAIR to CURE: Guidelines for Computational Models of Biological Systems, introduces the CURE principles—Credible, Understandable, Reproducible, and Extensible—as a framework to improve the development and use of computational models in biology and medicine. |
2025-02-12
König Lab at EP PerMed Conference on Personalised Medicine Research

|
We are excited to announce that Mariia Myshkina, Elisabetta Casabianca, and Matthias König participated in the EP PerMed Conference on Personalised Medicine Research, taking place on February 11–12, 2025, in Berlin, Germany. https://www.eppermed.eu/news-events/events/ep-permed-conference-on-personalised-medicine-research/ |
2025-01-20
Launch of New Research Internship in PBPK/PD Modeling
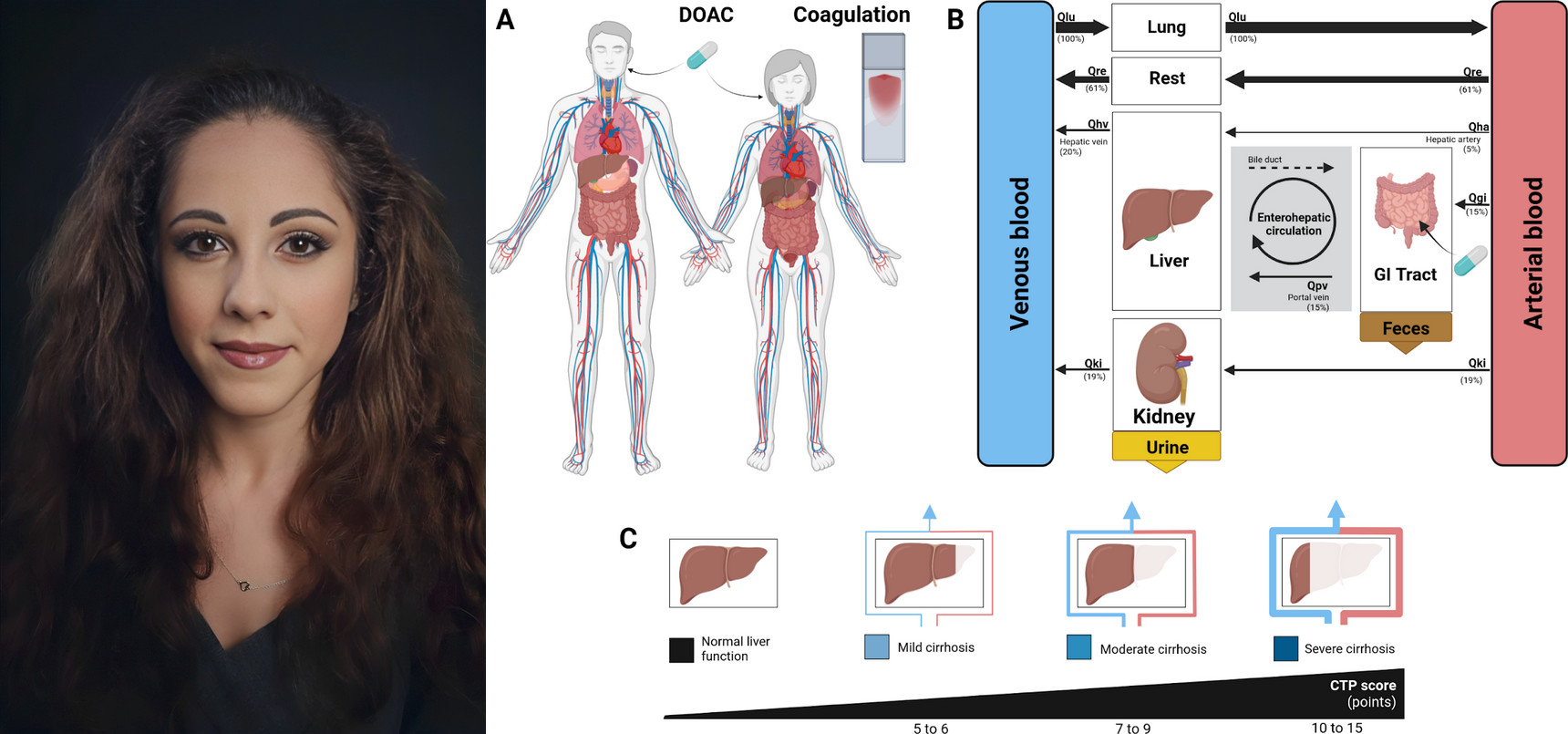
|
PBPK Model Development for Tirzepatide in Diabetes and Obesity Treatment |
2025-01-07
Launch of New Research Internship in PBPK/PD Modeling

|
PBPK Model Development for Tirzepatide in Diabetes and Obesity Treatment |
2024-12-20
Circle U. Funds AlgoNomy to Combat Digital Paternalism in AI-Driven Healthcare

|
Circle U. has funded AlgoNomy: Advancing Doctor-Patient Autonomy in AI-Driven Healthcare, a collaborative project tackling the issue of digital paternalism—where algorithms dominate healthcare decisions over human collaboration. Partnering with six leading European universities, AlgoNomy aims to develop ethical, transparent, and patient-centered AI systems that preserve autonomy and promote shared decision-making. |
2024-10-01
Humboldt Internship Program 2025 - Computational Modeling of Drug Detoxification - A Systems Medicine Approach

|
We offer Internships in 2025, academic three-month research stays at Humboldt-Universität zu Berlin. The application for the Summer Term Internships 2025 is open from 11 November to 20 December 2024. |
2024-08-30
Launch of Two New Research Projects in PBPK/PD Modeling
|
Ennie Tensil and Michelle Elias start Bachelor projects on losartan and glimeperide |
2024-08-28
Master Thesis Jonas Küttner
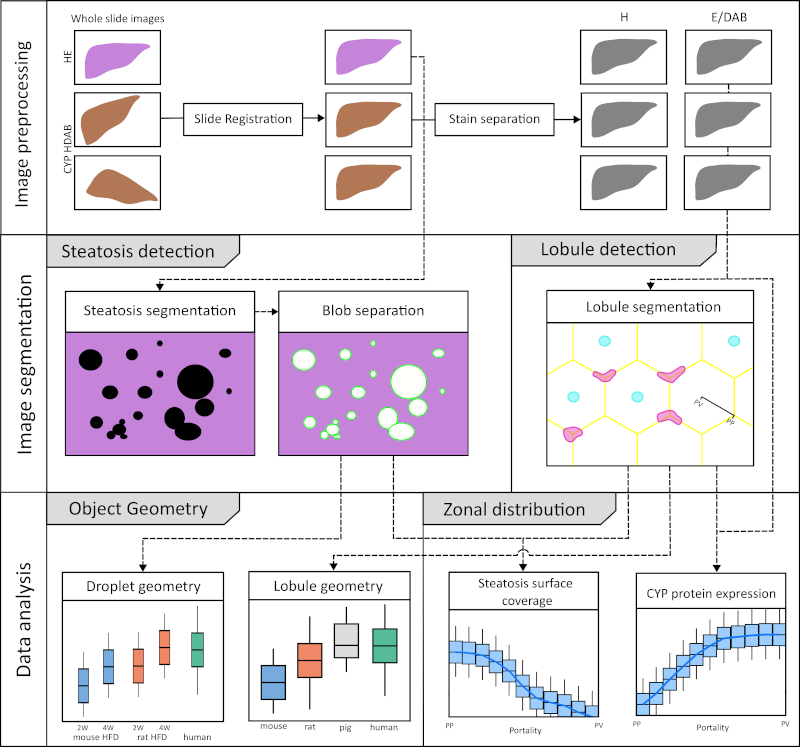
|
Jonas Küttner submitted his Master thesis developing a workflow for image analysis in the liver |
2024-08-08
Open Science Ambassador of the BUA

|
Proud to be selected as Open Science Ambassador of the Berlin University Alliance (BUA) in 2024. https://www.berlin-university-alliance.de/en/commitments/research-quality/open-science/index.html |
2024-07-18
Participation in DHPE

|
Matthias König selected for the Digital Health Professions Educator (DHPE) Program https://dsfz.charite.de/hochschuldidaktik/digital_health_professions_educator/ |
2024-07-18
Humboldt Internship Program (HIP) conference

|
Proud to participate in Humboldt Internship Program 2024. Great work by Vera, Deepa and Prerna. |
2024-07-15
Final ecosystem meeting of EDITH on building the VHT

|
Proud to participate in the final ecosystem meeting of EDITH on building the virtual human twin (VHT) in Amsterdam. |
2024-07-01
New project: Vera Tereshchuk starts SGLT2 PBPK Modeling Project
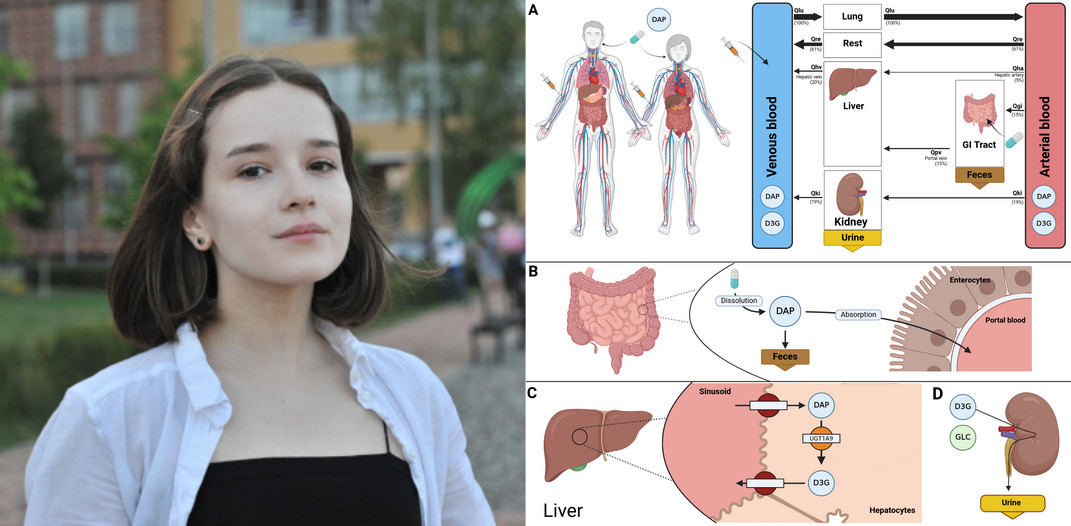
|
Vera Tereshchuk will develop a PBPK models for canagliflozin, aiming to enhance understanding of pharmacokinetics and pharmacodynamics in diabetes. |
2024-06-25
PAGE2024: Presentation of our research in Rome

|
Mariia Myshkina and Matthias König presented our captopril modeling results during the PAGE2024 (Population Approach Group Europe) meeting in Rome. |
2024-06-09
Bachelor Thesis Afruja Hossain
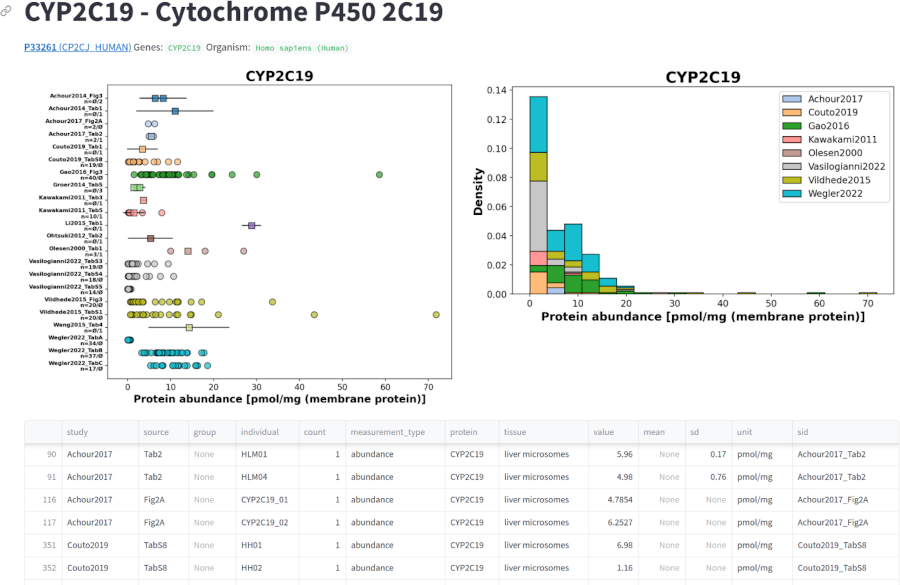
|
Afruja Hossain finished her Bachelor thesis developing a database of CYP and UGT enzymes |
2024-06-07
Successful Completion of One-Week Pharmacokinetic Modelling Course
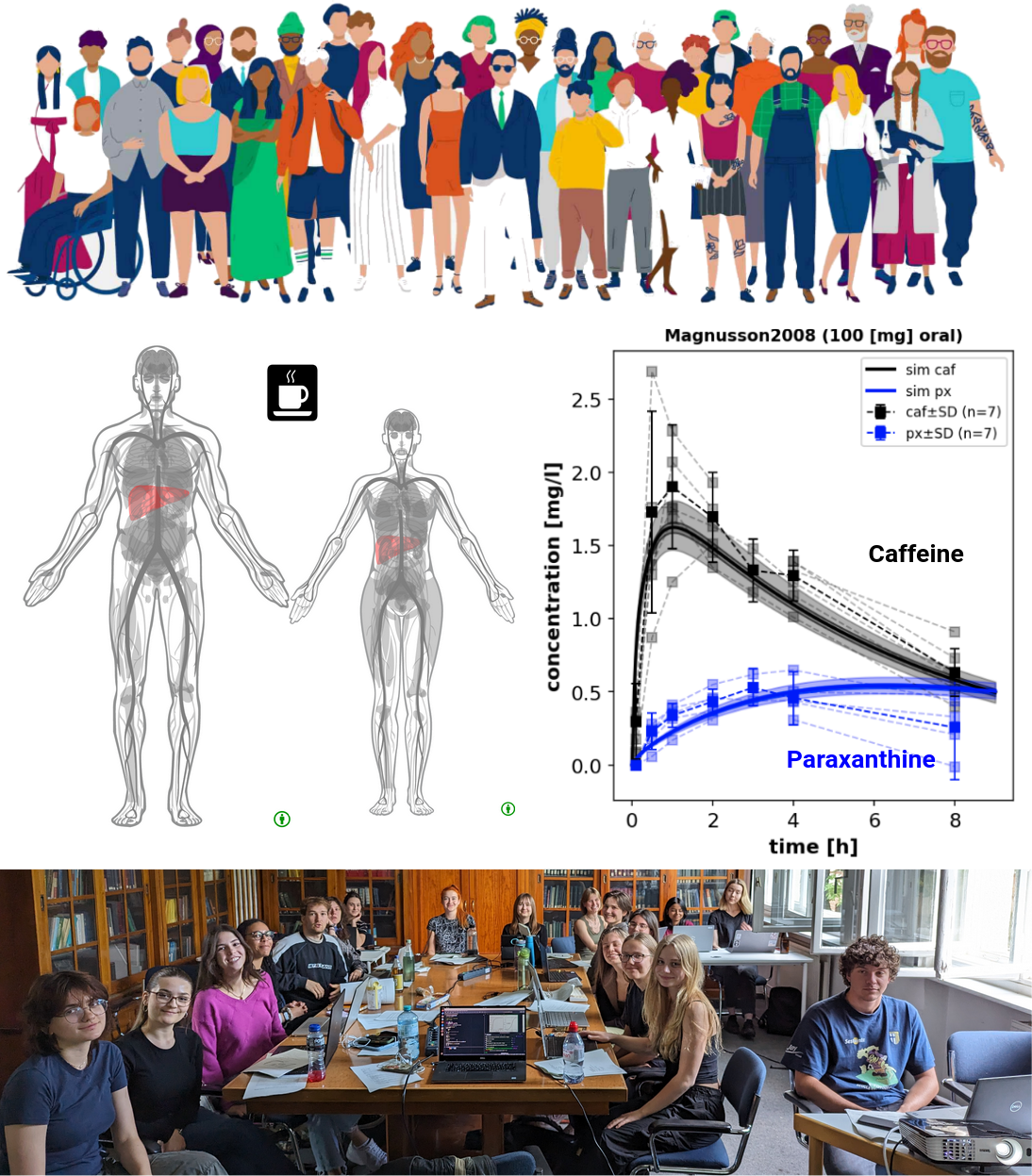
|
We are excited to announce the successful completion of our one-week intensive course on Pharmacokinetic Modelling, where students delved into principles, drug distribution, clearance, elimination, and various modelling techniques to enhance drug therapy and development. |
2024-05-23
New Project Launch via Humboldt Internship Program (HIP): PK-LLM by Prerna Parakkat
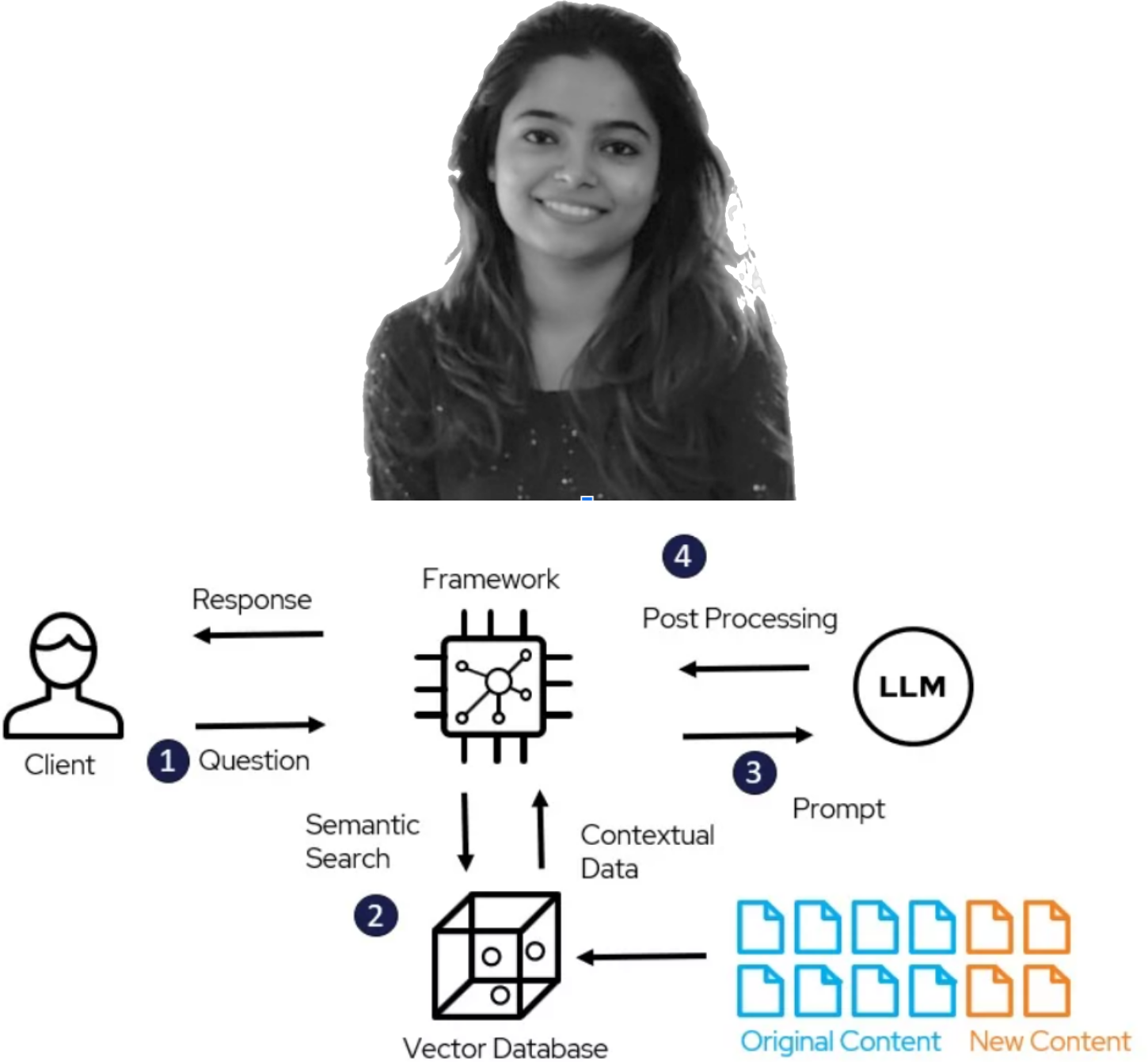
|
We are excited to announce the start of a new project under the Humboldt Internship Program (HIP) by Prerna Parakkat. Prerna will be working on PK-LLM: Implementing a Large Language Model (LLM) as an Expert System for Pharmacokinetic (PK) Data Curation from Scientific Publications. |
2024-05-16
Preprint: Cross-Species Variability in Lobular Geometry and Cytochrome P450 Hepatic Zonation: Insights into CYP1A2, CYP2E1, CYP2D6 and CYP3A4
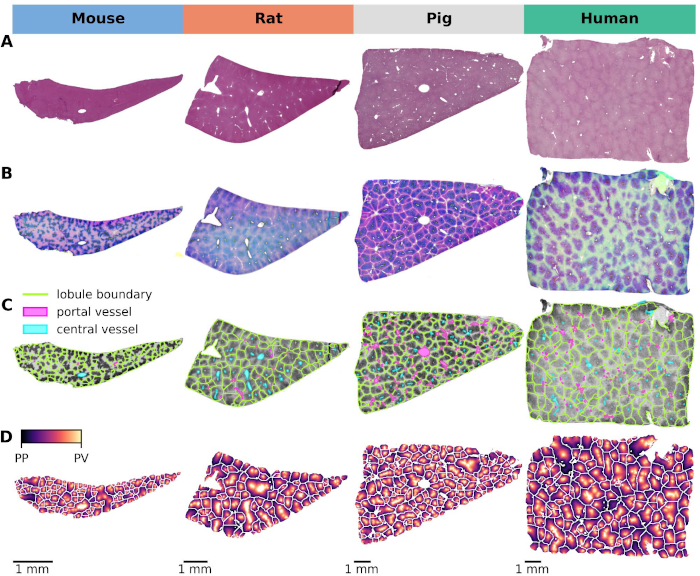
|
Our latest publication on 'Cross-Species Variability in Lobular Geometry & Cytochrome P450 Hepatic Zonation' reveals insights into liver function across species using WSI technology. https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2024.1404938/full |
2024-04-15
New projects: Nike Nemitz and Yusuf Ali Kulanoglu start PBPK Modeling Projects
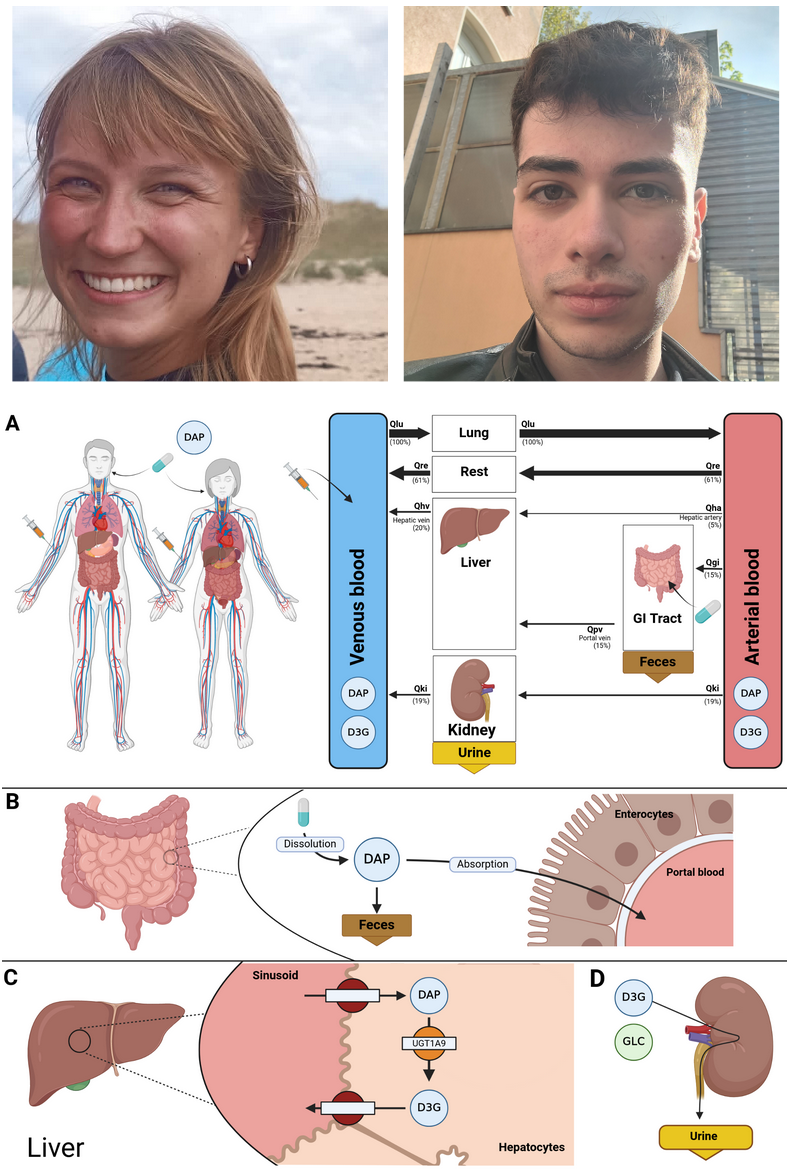
|
Nike Nemitz and Yusuf Ali Kulanoglu will develop PBPK models for dapagliflozin and aliskiren, respectively, aiming to enhance understanding of pharmacokinetics and pharmacodynamics in diabetes and cardiovascular treatments. |
2024-04-15
Publication: The Simulation Experiment Description Markup Language (SED-ML): Language Specification for Level 1 Version 5

|
Level 1 Version 5 of SED-ML enhances the ability to annotate, archive, share, and reproduce computational simulation experiments by allowing modelers to define tasks, model changes, ranges, and outputs using the Kinetic Simulation Algorithm Ontology (KiSAO). https://www.degruyter.com/document/doi/10.1515/jib-2024-0008/html |
2024-05-05
Master Thesis Shubhankar Palwankar
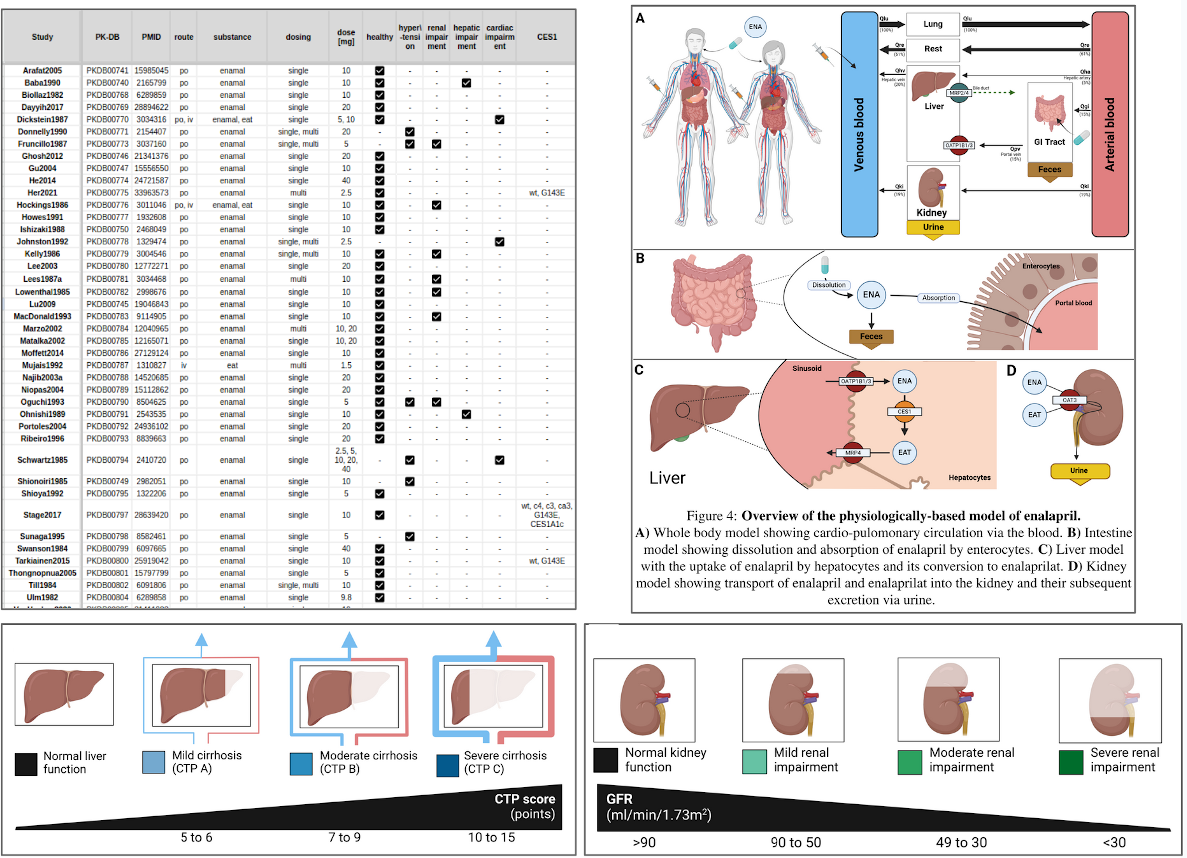
|
Shubhankar Palwankar defended his Master Thesis developing a PBPK model of enalapril |
2024-03-29
New multiscale liver model unveils zonation-function dynamics in Drug-Induced Liver Injury (DILI).

|
We're excited to announce the preprint of our latest research, 'Simulation of zonation-function relationships in the liver using coupled multiscale models: Application to drug-induced liver injury.' This study introduces a multiscale digital twin of the liver, integrating models from cellular to lobular scales, and sheds light on how liver zonation patterns influence drug-induced liver injury. |
2024-03-01
PBPK Model of Morphine Project
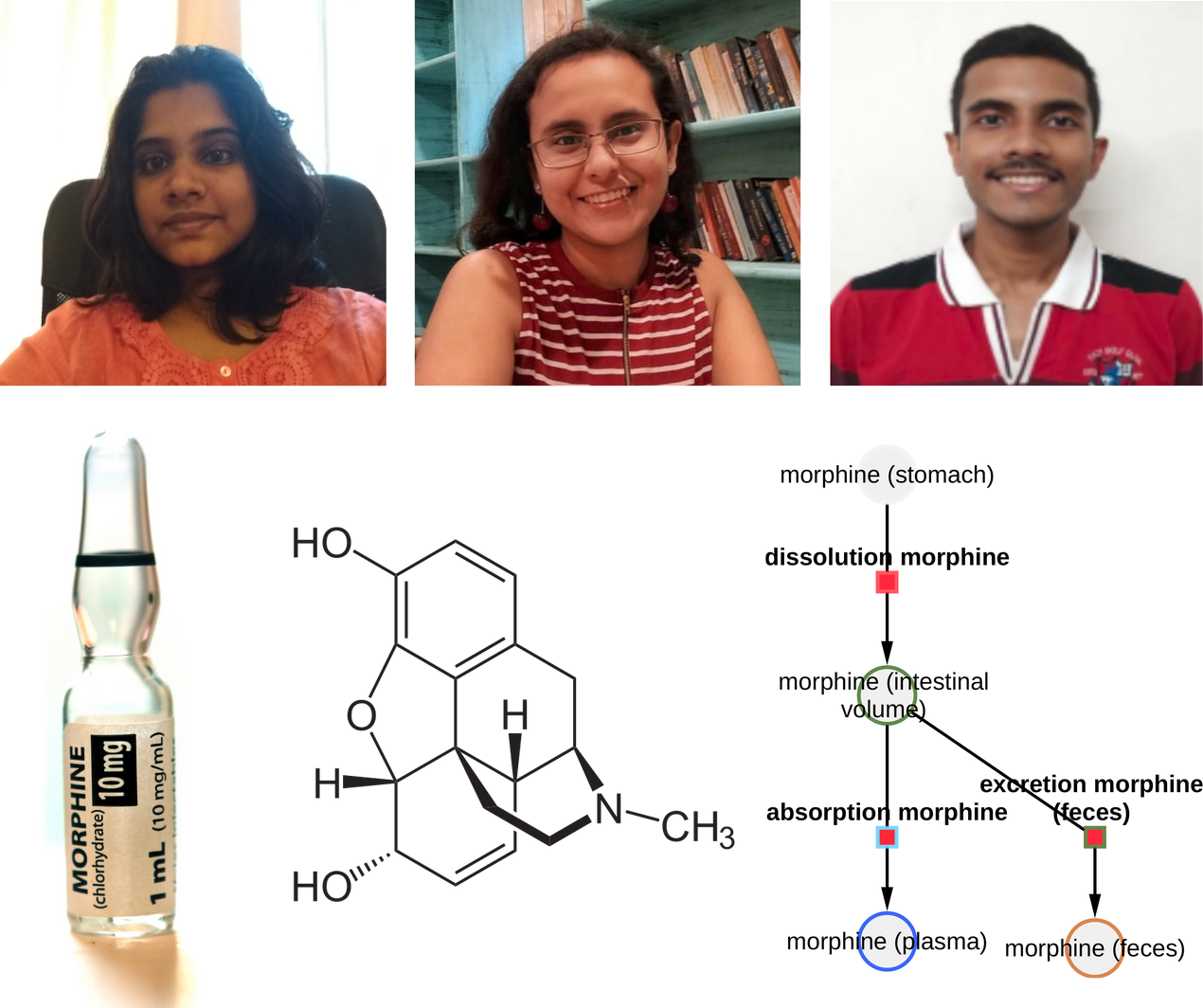
|
New Internship Project Launch: Development of a Physiologically Based Pharmacokinetics Model of Morphine. Deepa supported by Rohini and Rohit will develop a PBPK model of morphine. |
2024-02-01
Mariia Myshkina has started her PhD in the ATLAS project.

|
We welcome Mariia to our team who will be working on computational modeling of the liver within ATLAS. |
2024-01-18
Participation in the First EDITH-CSA Ecosystem Meeting - Advancing the Virtual Human Twin

|
We participated in the first EDITH-CSA 'Ecosystem meeting on building the Virtual Human Twin was held at the École Polytechnique de Paris on January 18 and 19, 2024. |
2023-12-29
Preprint: Cross-Species Variability in Lobular Geometry and Cytochrome P450 Hepatic Zonation: Insights into CYP1A2, CYP2E1, CYP2D6 and CYP3A4

|
Our latest preprint on 'Cross-Species Variability in Lobular Geometry & Cytochrome P450 Hepatic Zonation' reveals insights into liver function across species using WSI technology. |
2023-11-03
Eleven Strategies for Making Reproducible Research and Open Science Training the Norm at Research Institutions

|
We are pleased to announce our latest paper in eLife that provides a roadmap for integrating reproducible research and open science practices into academic work. |
2023-11-03
Your Dietary Digitial Twin (DDtwin)

|
Advancing Precision Nutrition: Workshop Success on Dietary Digital Twin, a workshop in Leiden from 30 October - 3 November 2023. https://www.lorentzcenter.nl/your-dietary-digital-twin-ddtwin.html |
2023-10-31
Humboldt Internship Program 2024 - Computational Modeling of Drug Detoxification - A Systems Medicine Approach

|
We offer Internships in 2024, academic three-month research stays at Humboldt-Universität zu Berlin. The application for the Summer Term Internships 2024 is open from 03 November to 10 December 2023. |
2023-10-10
e:Med Meeting 2023 on Systems Medicine: Quantifying Fat Zonation in Liver Lobules: An Integrated Multiscale In-silico Model

|
We successfully presented our research on reproducible digital twins for personalized liver function assessment at the e:Med Meeting 2023 on Systems Medicine. |
2023-09-18
Preprint: Quantifying Fat Zonation in Liver Lobules: An Integrated Multiscale In-silico Model
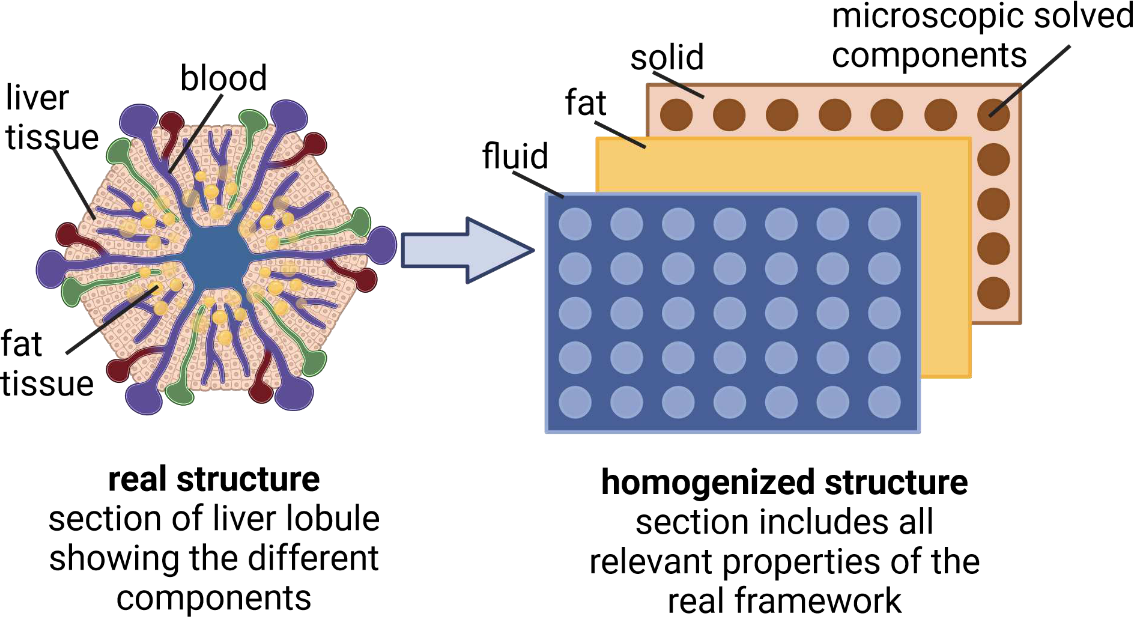
|
Our latest preprint is now out: An integrated multiscale model for quantifying fat zonation in liver lobules. |
2023-08-28
Hannah Menghis from Brown University starts her internship

|
We welcome Hannah Menghis who will establish a PBPK model of semaglutide. |
2023-08-02
A pathway model of glucose-stimulated insulin secretion in the pancreatic β-cell
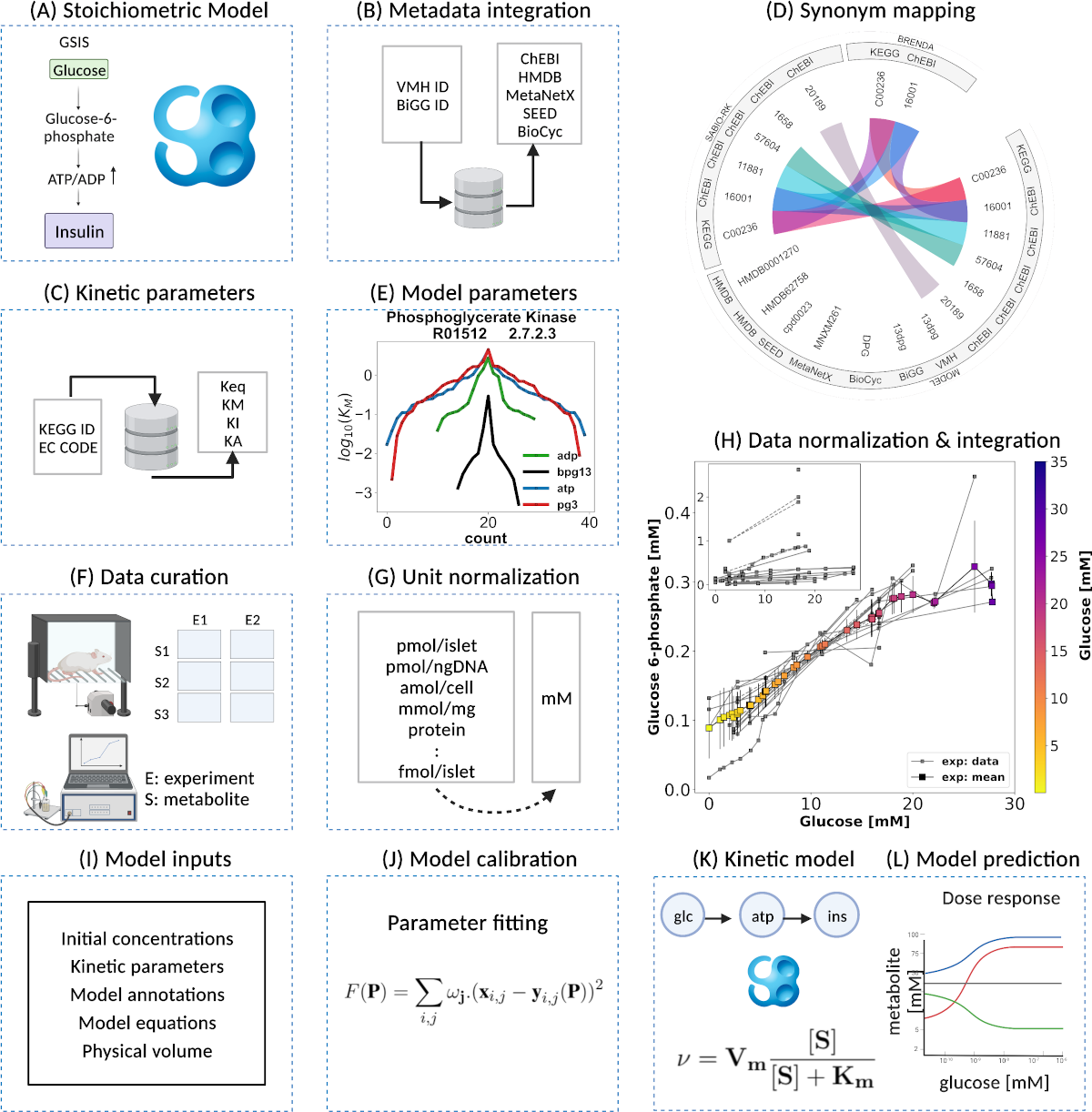
|
Our latest publication is now on Frontiers of Endocrinology: A pathway model of glucose-stimulated insulin secretion in the pancreatic β-cell |
2023-07-17
Bachelor Thesis Beatrice Stemmer Mallol
|
Beatrice Stemmer Mallol submitted her Bachelor thesis developing a PBPK model of talinolol |
2023-07-07
PhD Defense of Jan Grzegorzewski
|
Jan Grzegorzewski defended his PhD thesis on physiologically based pharmacokinetic modeling (PBPK) for dynamical liver function tests and Cytochrome P450 (CYP) phenotyping. |
2023-07-01
Xenia Petukhova and Zahra Bahri start Humboldt Internship Program

|
Humboldt Internship Program 2023 - Computational Modeling of Drug Detoxification - A Systems Medicine Approach |
2023-06-16
Keynote: Advancing Liver Function Assessment: Personalized and Stratified Approaches with Standardized Computational Models and Data

|
Keynote talk at the Workshop on Computational Models in Biology and Medicine 2023 organized by Nicole Radde and Sebastian Höpfl. https://docs.google.com/presentation/d/1hi7-4bwzqFJYiYQkbSBHNQivSPA5T3YHB9OLZqJG0rA/edit?usp=sharing |
2023-05-28
Preprint: Eleven Strategies for Making Reproducible Research and Open Science Training the Norm at Research Institutions
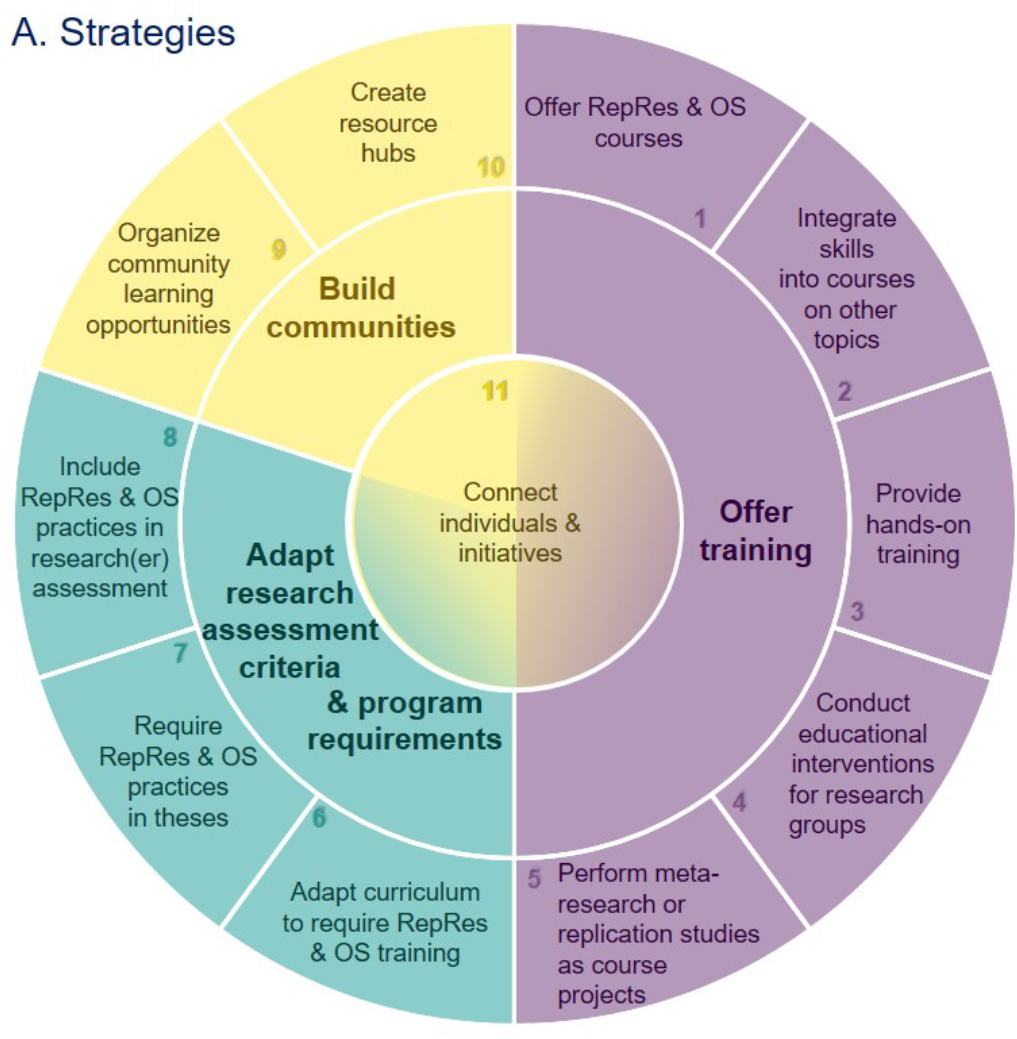
|
We present 11 strategies for making #OpenScience & #ReproducibleResearch the norm at research institutions, with tips for implementation & resources. Let's join hands to make these practices standard in the scientific community! |
2023-04-16
Preprint: A physiologically based pharmacokinetic model for CYP2E1 phenotyping via chlorzoxazone
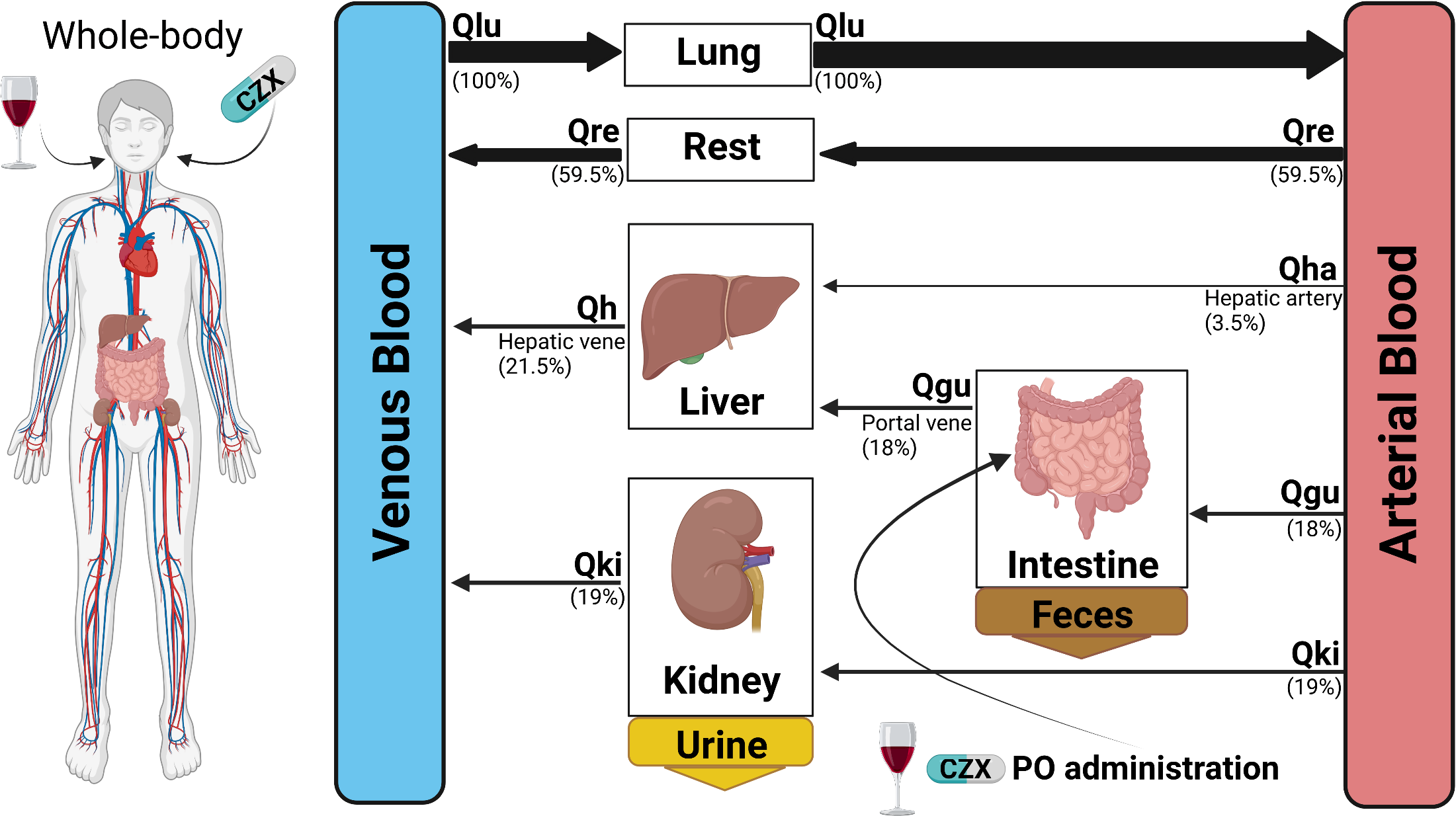
|
Our latest preprint is now on bioRxiv: A physiologically based pharmacokinetic model for CYP2E1 phenotyping via chlorzoxazone |
2023-04-03
Michael Stifel Price to promote interdisciplinary, data-driven research

|
M. Albadry, S. Höpfl, and M. König won the QuaLiPerF Michael Stifel Price for interdisciplinary, data-driven research. |
2023-03-29
Publication: Specifications of standards in systems and synthetic biology: status and developments in 2022 and the COMBINE meeting 2022
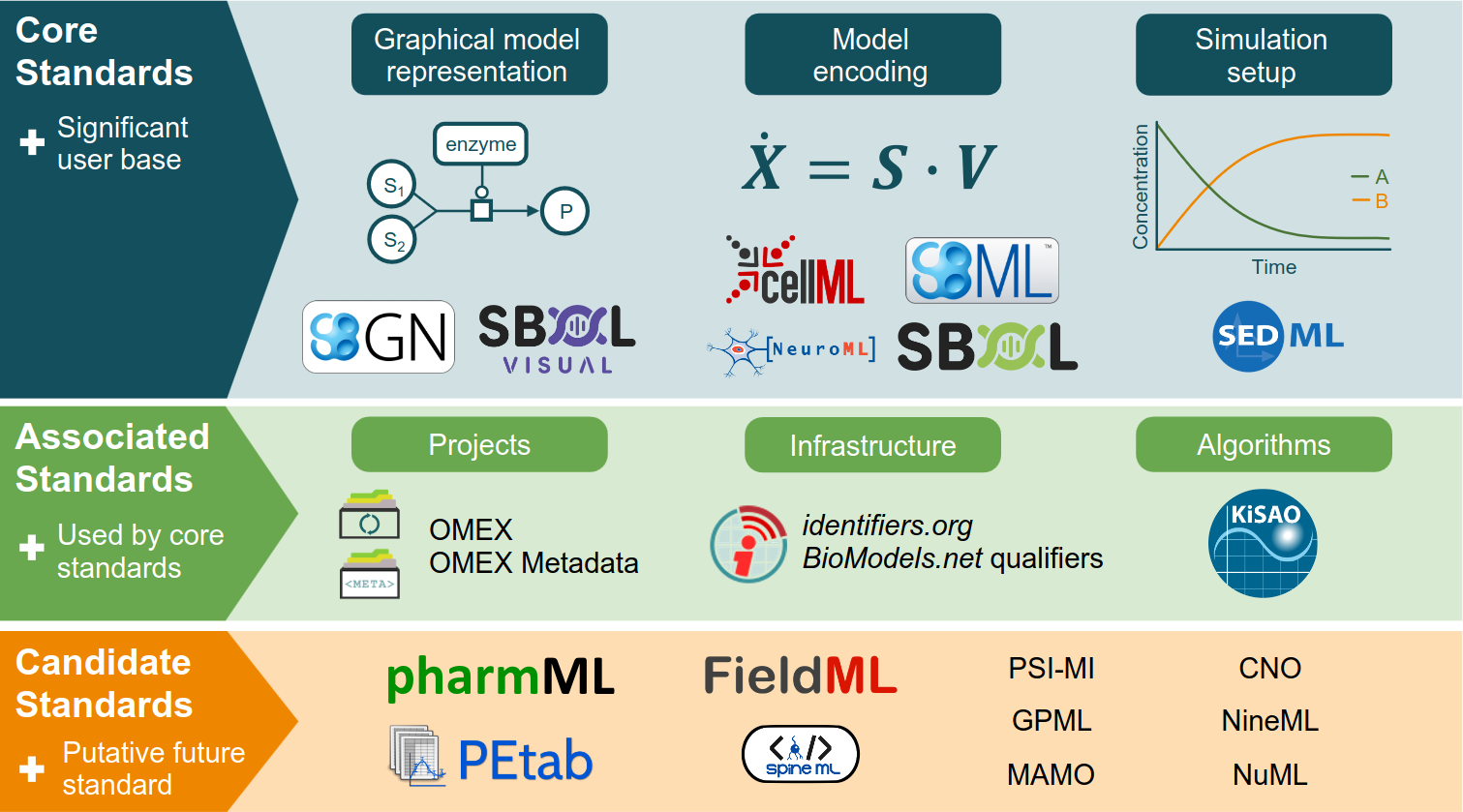
|
Our overview of specifications in systems and synthetic biology with a brief overview of the COMBINE 2022 is available. |
2023-03-13
Preprint: A consensus model of glucose-stimulated insulin secretion in the pancreatic beta-cell

|
Our latest preprint is now on bioRxiv: A consensus model of glucose-stimulated insulin secretion in the pancreatic beta-cell |
2023-02-12
PBPK modelling of sorafenib and Gd-EOB-DTPA

|
Frances Okibedi and Narudeem Sojimade will develop models of the anticancer drug sorafenib in HCC and the liver contrast agent Gd-EOB-DTPA. |
2023-02-12
X-Research Group - Digital twins for the treatment of hypertension
|
M. König receives funding of the Berlin University Alliance and the DFG for the X-Student Research Group: Digital twins for the treatment of hypertension |
2023-02-07
M. König has been elected as PEtab editor (2023-2026)

|
Proud to be elected editor for PEtab, a data format for specifying parameter estimation problems in systems biology. |
2023-02-03
Preprint: Simvastatin therapy in different subtypes of hypercholesterolemia
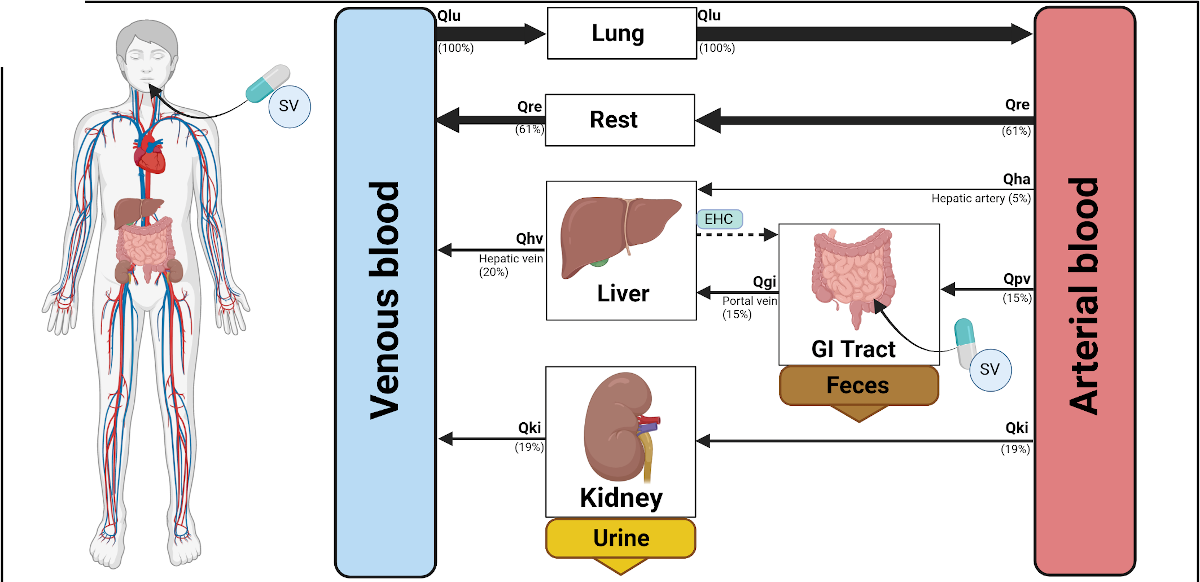
|
Our latest preprint is now on medRxiv: Simvastatin therapy in different subtypes of hypercholesterolemia - a physiologically based modelling approach. https://www.medrxiv.org/content/10.1101/2023.02.01.23285358v1 |
2023-02-01
ATLAS - AI and Simulation for Tumor Liver ASsessment
|
The BMBF is funding our joint project ATLAS in the "Computational Life Sciences - AI Methods for Systems Medicine". |
2022-11-17
New Bachelor Student - Afruja Hossain

|
Afruja Hossain started working in our lab on a systematic overview of the protein variability of cytochrome P450 (CYP450) isoforms in the human liver. |
2022-11-15
Visiting EU parliament and attending STOA AI session

|
We were selected to participate in an European policy journey on the topic EU research funding and science communication by the Berlin University Alliance (BUA). https://www.berlin-university-alliance.de/news/items/2022/221017-bruessel-reise.html |
2022-10-26
Humboldt Internship Program 2023 - Computational Modeling of Drug Detoxification - A Systems Medicine Approach

|
We offer Internships in 2023, application open from 01 Nov - 09 Dec 2022. |
2022-10-24
Dextromethorphan publication
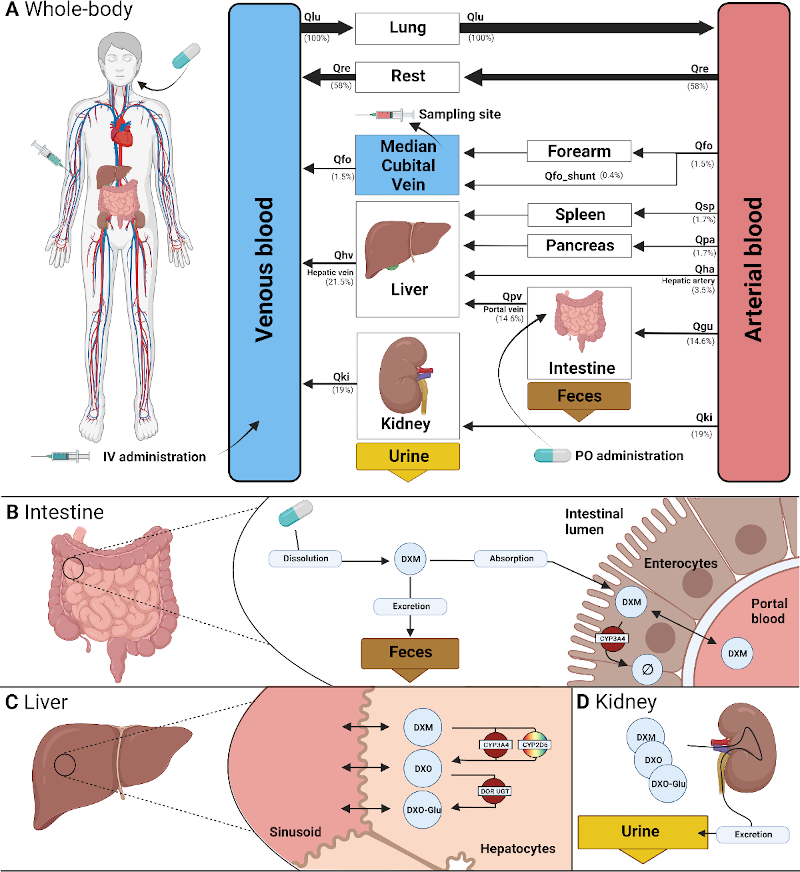
|
How does CYP2D6 genotype effect metabolic phenotyping? See our latest paper in Frontiers in Pharmacology for answers. https://www.frontiersin.org/articles/10.3389/fphar.2022.1029073/full |
2022-10-24
X-Research Group

|
M. König receives funding of the Berlin University Alliance and the DFG for the X-Student Research Group: Physiologically based modeling of drugs: ACE inhibitors |
2022-10-06
COMBINE 2022


|
We organized the Computational Modeling in Biology Network (COMBINE) meeting in Berlin this year with > 100 participants from 06-08 October 2022. |
2022-10-10
ICSB 2022 - Presentation and Posters
|
We could present our work on physiologically based pharmacokinetics (PBPK) modeling at ICSB2022. |