Python library for SBGN
Matthias König
Humboldt-University Berlin, Faculty of Life Science, Systems Medicine of Liver
https://livermetabolism.com
2025-04-15
SBGN
- Systems Biology Graphical Notation
- Standardise graphical notation used in maps of biological processes
- Process Description (PD): temporal courses of biochemical interactions in a network
- Entity Relationship (ER): all relationships between participates, regardless of the temporal aspects
- Activity Flow (AF): flow of information between biochemical entities in a network

libsbgnpy
- Python library for SBGN
- Supports AF, PD, ER
- Supports libsbgn/0.3
- new: support for render extension
- new: python dataclasses with type annotations
- new: py3.12, py3.13 support
- new: more and better documentation: https://matthiaskoenig.github.io/libsbgnpy/

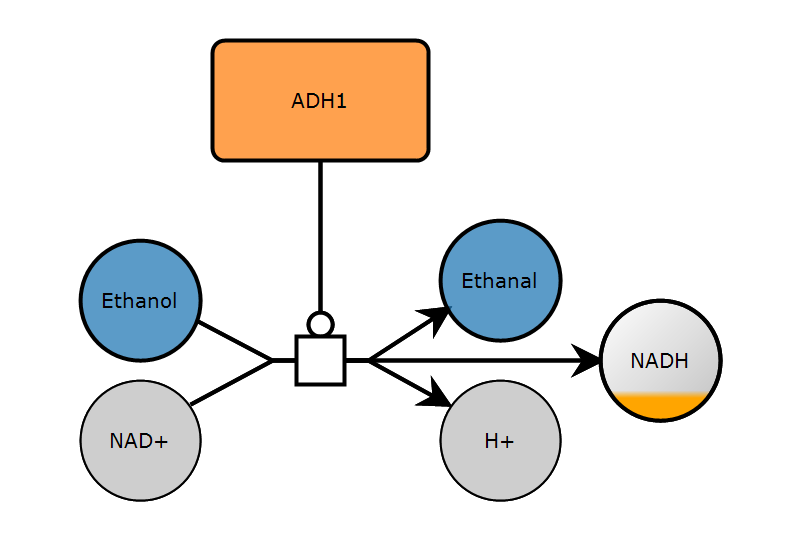
1. Create map
2. Add glyphs
"""Ethanol example."""
map.glyph.extend([
Glyph(
class_value=GlyphClass.SIMPLE_CHEMICAL,
id="ethanol",
label=Label(text="Ethanol"),
bbox=Bbox(x=40, y=120, w=60, h=60),
),
Glyph(
class_value=GlyphClass.SIMPLE_CHEMICAL,
id="ethanal",
label=Label(text="Ethanal"),
bbox=Bbox(x=220, y=110, w=60, h=60),
),
Glyph(
class_value=GlyphClass.MACROMOLECULE,
id="adh1",
label=Label(text="ADH1"),
bbox=Bbox(x=106, y=20, w=108, h=60),
),
Glyph(
class_value=GlyphClass.SIMPLE_CHEMICAL,
id="h",
label=Label(text="H+"),
bbox=Bbox(x=220, y=190, w=60, h=60),
clone=Glyph.Clone(),
),
Glyph(
class_value=GlyphClass.SIMPLE_CHEMICAL,
id="nad",
label=Label(text="NAD+"),
bbox=Bbox(x=40, y=190, w=60, h=60),
clone=Glyph.Clone(),
),
Glyph(
class_value=GlyphClass.SIMPLE_CHEMICAL,
id="glyph_nadh",
label=Label(text="NADH"),
bbox=Bbox(x=300, y=150, w=60, h=60),
clone=Glyph.Clone(),
),
# glyph with ports (process)
Glyph(
class_value=GlyphClass.PROCESS,
id="pn1",
orientation=GlyphOrientation.HORIZONTAL,
bbox=Bbox(x=148, y=168, w=24, h=24),
port=[
Port(x=136, y=180, id="pn1.1"),
Port(x=184, y=180, id="pn1.2"),
],
),
])2. Add glyphs
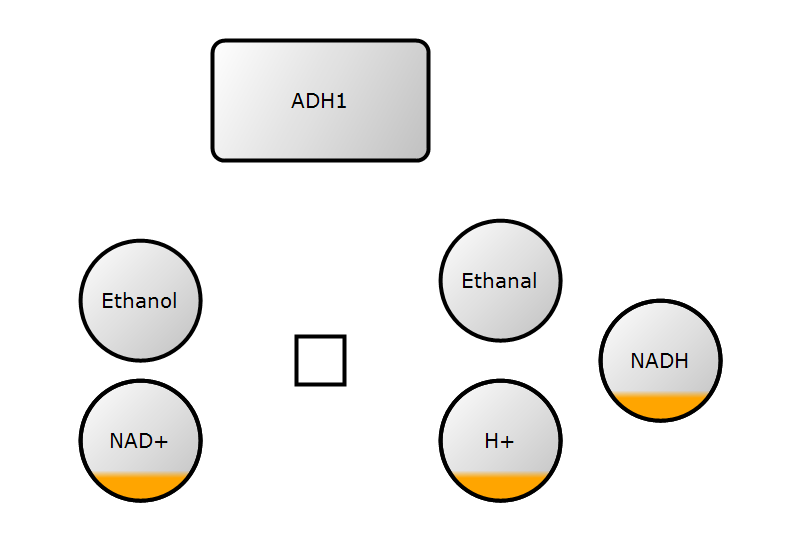
3. Add arcs
"""Ethanol example."""
# create arcs and set the start and end points
map.arc.extend(
[
Arc(
id="a01",
class_value=ArcClass.CONSUMPTION,
source="ethanol",
target="pn1.1",
start=Arc.Start(x=98, y=160),
end=Arc.End(x=136, y=180),
),
Arc(
id="a02",
class_value=ArcClass.PRODUCTION,
source="pn1.2",
target="nadh",
start=Arc.Start(x=184, y=180),
end=Arc.End(x=300, y=180),
),
Arc(
id="a03",
class_value=ArcClass.CATALYSIS,
source="adh1",
target="pn1",
start=Arc.Start(x=160, y=80),
end=Arc.End(x=160, y=168),
),
Arc(
id="a04",
class_value=ArcClass.PRODUCTION,
source="pn1.2",
target="h",
start=Arc.Start(x=184, y=180),
end=Arc.End(x=224, y=202),
),
Arc(
id="a05",
class_value=ArcClass.PRODUCTION,
source="pn1.2",
target="ethanal",
start=Arc.Start(x=184, y=180),
end=Arc.End(x=224, y=154),
),
Arc(
id="a06",
class_value=ArcClass.CONSUMPTION,
source="nad",
target="pn1.1",
start=Arc.Start(x=95, y=202),
end=Arc.End(x=136, y=180),
),
]
)3. Add arcs
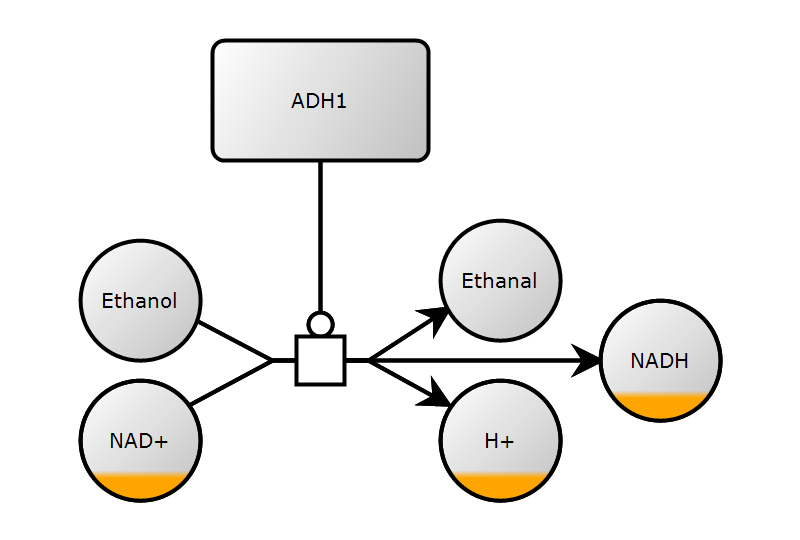
4. Add render information
render_info = RenderInformation(
id="ethanol_render_info",
program_name="libsbgnpy",
program_version="0.4.0",
list_of_color_definitions=ListOfColorDefinitions(
color_definition=[
ColorDefinition(id="blue", value="#1f77b4bb"),
ColorDefinition(id="orange", value="#ff7f0ebb"),
ColorDefinition(id="white", value="#000000"),
ColorDefinition(id="grey", value="#cccccccc"),
ColorDefinition(id="black", value="#ffffff"),
]
),
list_of_gradient_definitions=ListOfGradientDefinitions(),
list_of_styles=ListOfStyles(
[
Style(
id_list="ethanol ethanal",
g=G(stroke="black", stroke_width=2, fill="blue"),
),
Style(
id_list="adh1",
g=G(stroke="black", stroke_width=2, fill="orange"),
),
Style(
id_list="nad nadh h",
g=G(stroke="black", stroke_width=1, fill="grey"),
),
]
),
)5. Save and render
6. Use other tools in the workflow
SBGN maps can be further modified in additional tools such as Newt: Ethanol Example Newt

Want to know more