At the König Lab, we merge the fields of Systems Medicine and Systems Biology to delve deep into drug
metabolism and liver function. By harnessing the power of computational modeling, data science,
bioinformatics analysis, and machine learning, we analyze and interpret datasets spanning biological,
medical, and clinical spheres.
A hallmark of our research is the generation of predictive digital twins mirroring individual human
physiology. These models are instrumental in shedding light on inter-individual variances, paving the way
for personalized treatments—from tailoring drug dosages to understanding the nuanced impacts of lifestyle on
drug metabolism. For more information, please see our
publications and projects.
We are actively involved in the standardization and reproducibility efforts in Systems Biology and Systems
Medicine.
We are actively involved in the standardization and reproducibility efforts in Systems Biology and Systems
Medicine.
For more information, see our
| 2024 |

|
Simulation of zonation-function relationships in the liver using coupled multiscale models: Application to drug-induced liver injury
Steffen Gerhäusser, Lena Lambers, Luis Mandl, Julian Franquinet, Tim Ricken, Matthias König
bioRxiv 2024.03.26.586870; (preprint). doi:10.1101/2024.03.26.586870
Multiscale modeling requires the coupling of models on different scales, often based on different mathematical approaches and developed by different research teams. This poses many challenges, such as defining interfaces for coupling, reproducible exchange of submodels, efficient simulation of the models, or reproducibility of results. Here, we present a multiscale digital twin of the liver that couples a partial differential equation (PDE)-based porous media approach for the hepatic lobule with cellular-scale ordinary differential equation (ODE)-based models. The models based on the theory of porous media describe transport, tissue mechanical properties, and deformations at the lobular scale, while the cellular models describe hepatic metabolism in terms of drug metabolism and damage in terms of necrosis. The resulting multiscale model of the liver was used to simulate perfusion-zonation-function relationships in the liver spanning scales from single cell to the lobulus. The model was applied to study the effects of (i) protein zonation patterns (metabolic zonation) and (ii) drug concentration dependence on spatially heterogeneous liver damage in the form of necrosis. Depending on the zonation pattern, different liver damage patterns could be reproduced, including periportal and pericentral necrosis as seen in drug-induced liver injury (DILI). Increasing the drug concentration led to an increase in the observed damage pattern. A key point for the success was the integration of domain-specific simulators based on standard exchange formats, i.e., libroadrunner for the high-performance simulation of ODE-based systems and FEBio for the simulation of the continuum-biomechanical part. This allows a standardized and reproducible exchange of cellular scale models in the Systems Biology Markup Language (SBML) between research groups.
Keywords:
liver,
computational model,
biomechanics,
porous media,
drug metabolism,
digital twin,
DILI,
cytochrome P450,
|
|
| 2024 |

|
Quantifying fat zonation in liver lobules: an integrated multiscale in silico model combining disturbed microperfusion and fat metabolism via a continuum biomechanical bi-scale, tri-phasic approach
Lena Lambers, Navina Waschinsky, Jana Schleicher, Matthias König, Hans-Michael Tautenhahn, Mohamed Albadry, Uta Dahmen and Tim Ricken
Biomech Model Mechanobiol. 2024 Feb 25.. doi:10.1007/s10237-023-01797-0. pmid:38402347
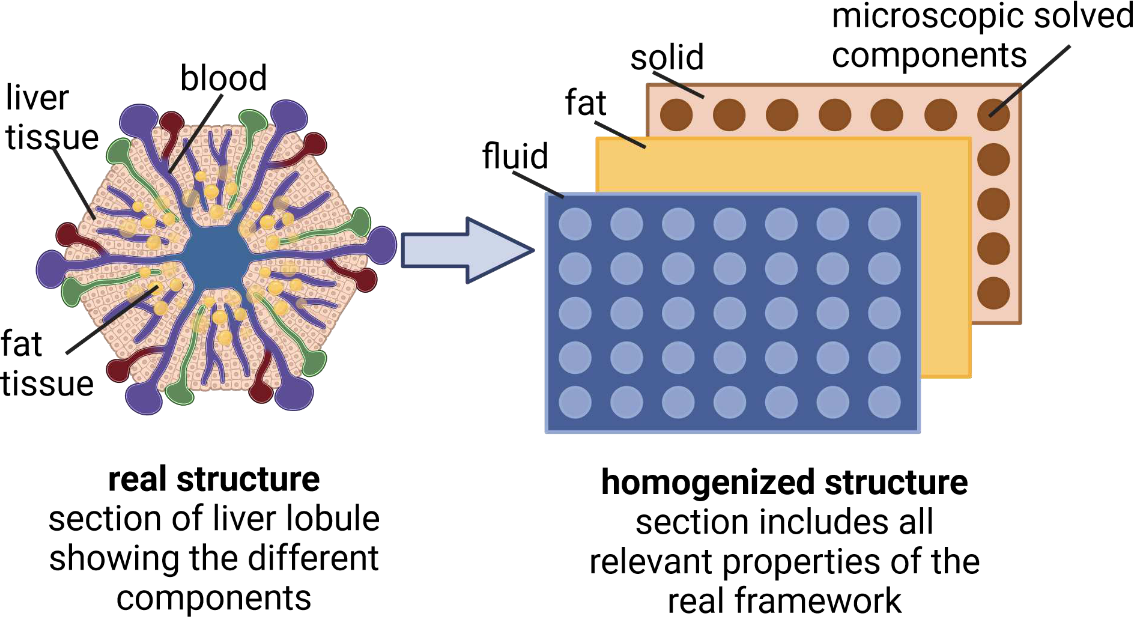
Metabolic zonation refers to the spatial separation of metabolic functions along the sinusoidal axes of the liver. This phenomenon forms the foundation for adjusting hepatic metabolism to physiological requirements in health and disease (e.g., metabolic dysfunction-associated steatotic liver disease/MASLD). Zonated metabolic functions are influenced by zonal morphological abnormalities in the liver, such as periportal fibrosis and pericentral steatosis. We aim to analyze the interplay between microperfusion, oxygen gradient, fat metabolism and resulting zonated fat accumulation in a liver lobule. Therefore we developed a continuum biomechanical, tri-phasic, bi-scale, and multicomponent in silico model, which allows to numerically simulate coupled perfusion-function-growth interactions two-dimensionally in liver lobules. The developed homogenized model has the following specifications: (i) thermodynamically consistent, (ii) tri-phase model (tissue, fat, blood), (iii) penta-substances (glycogen, glucose, lactate, FFA, and oxygen), and (iv) bi-scale approach (lobule, cell). Our presented in silico model accounts for the mutual coupling between spatial and time-dependent liver perfusion, metabolic pathways and fat accumulation. The model thus allows the prediction of fat development in the liver lobule, depending on perfusion, oxygen and plasma concentration of free fatty acids (FFA), oxidative processes, the synthesis and the secretion of triglycerides (TGs). The use of a bi-scale approach allows in addition to focus on scale bridging processes. Thus, we will investigate how changes at the cellular scale affect perfusion at the lobular scale and vice versa. This allows to predict the zonation of fat distribution (periportal or pericentral) depending on initial conditions, as well as external and internal boundary value conditions.
Keywords:
Multiscale Approach,
Homogenization,
Theory of Porous Media (TPM),
Growth Kinematics,
Metabolic dysfunction-Associated Steatotic Liver Disease (MASLD),
FEM boundary problem,
|
|
| 2024 |
|
Bayesian Modeling of Time Series Data (BayModTS) - A FAIR Workflow to Process Sparse and Highly Variable Data
Sebastian Höpfl, Mohamed Albadry, Uta Dahmen, Karl-Heinz Herrmann, Eva Marie Kindler, Matthias König, Jürgen Rainer Reichenbach, Hans-Michael Tautenhahn, Weiwei Wei, Wan-Ting Zhao, Nicole Erika Radde
(submitted)
Motivation. Systems biology aims to better understand living systems through mathematical modelling of experimental and clinical data. A pervasive challenge in quantitative dynamical modelling is the integration of time series measurements, which often have high variability and low sampling resolution. Approaches are required to utilise such information while consistently handling uncertainties. Results. We present BayModTS (Bayesian Modeling of Time Series data), a new FAIR (Findable, Accessible, Interoperable and Reusable) workflow for processing and analysing sparse and highly variable time series data. BayModTS consistently transfers uncertainties from data to model predictions, including process knowledge via parameterised models. Further, credible differences in the dynamics of different conditions can be identified by filtering noise. To demonstrate the power and versatility of BayModTS, we applied it to three hepatic datasets gathered from three different species and with different measurement techniques: (i) blood perfusion measurements by magnetic resonance imaging in rat livers after portal vein ligation, (ii) CT-based volumetric assessment of human liver remnants after clinical liver resection, and (iii) pharmacokinetic time series of different drugs in normal and steatotic mice. Availability and Implementation. The BayModTS codebase is available on GitHub at https://github.com/Systems-Theory-in-Systems-Biology/BayModTS. The repository contains a Python script for the executable BayModTS workflow and a widely applicable SBML (Systems Biology Markup Language) model for retarded transient functions. In addition, all examples from the paper are included in the repository. Data and code of the application examples are stored on DaRUS https://doi.org/10.18419/darus-3876.
Keywords:
Time Series Data,
Bayesian Inference,
Posterior Predictive Distribution,
SBML,
FAIR Principles,
Hepatology,
|
|
| 2024 |

|
SimLivA – Modelling ischemia reperfusion injury in the liver: A first step towards a clinical decision support tool.
Hans-Michael Tautenhahn, Tim Ricken, Uta Dahmen, Luis Mandl, Laura Bütow, Steffen Gerhäusser, Lena Lambers, Xinpei Chen, Elina Lehmann, Olaf Dirsch, Matthias König
GAMM-Mitteilungen. (2024), e202370003. doi:10.1002/gamm.202370003
The SIMulation supported LIVer Assessment for donor organs (SimLivA) project aims to develop a mathematical model to accurately simulate the influence of mechanical alterations in marginal liver grafts (specifically steatotic ones) and cold ischemia on early ischemia-reperfusion injury (IRI) during liver transplantation. Our project tackles significant research challenges, including the co-development of computational methodologies, experimental studies, clinical processes, and technical workflows. We aim to refine a continuum-biomechanical model for enhanced IRI prediction, collect pivotal experimental and clinical data, and assess the clinical applicability of our model. Our efforts involve augmenting and tailoring a coupled continuum-biomechanical, multiphase, and multi-scale partial differential equation-ordinary differential equation (PDE-ODE) model of the liver lobule, allowing us to numerically simulate IRI depending on the degree of steatosis and the duration of ischemia. The envisaged model will intertwine the structure, perfusion, and function of the liver, serving as a crucial aid in clinical decision-making processes. We view this as the initial step towards an in-silico clinical decision support tool aimed at enhancing the outcomes of liver transplantation. In this paper, we provide an overview of the SimLivA project and our preliminary findings, which include: a cellular model that delineates critical processes in the context of IRI during transplantation; and the integration of this model into a multi-scale PDE-ODE model using a homogenized, multi-scale, multi-component approach within the Theory of Porous Media (TPM) framework. The model has successfully simulated the interconnected relationship between structure, perfusion, and function—all of which are integral to IRI. Initial results show simulations at the cellular scale that describe critical processes related to IRI during transplantation. After integrating this model into a multiscale PDE-ODE model, first simulations were performed on the spatial distribution of key functions during warm and cold ischaemia. In addition, we were able to study the effect of tissue perfusion and temperature, two critical parameters in the context of liver transplantation and IRI.
Keywords:
ischemia-reperfusion injury,
IRI,
liver transplantation,
ODE,
PDE,
porous media,
|
|
| 2024 |


|
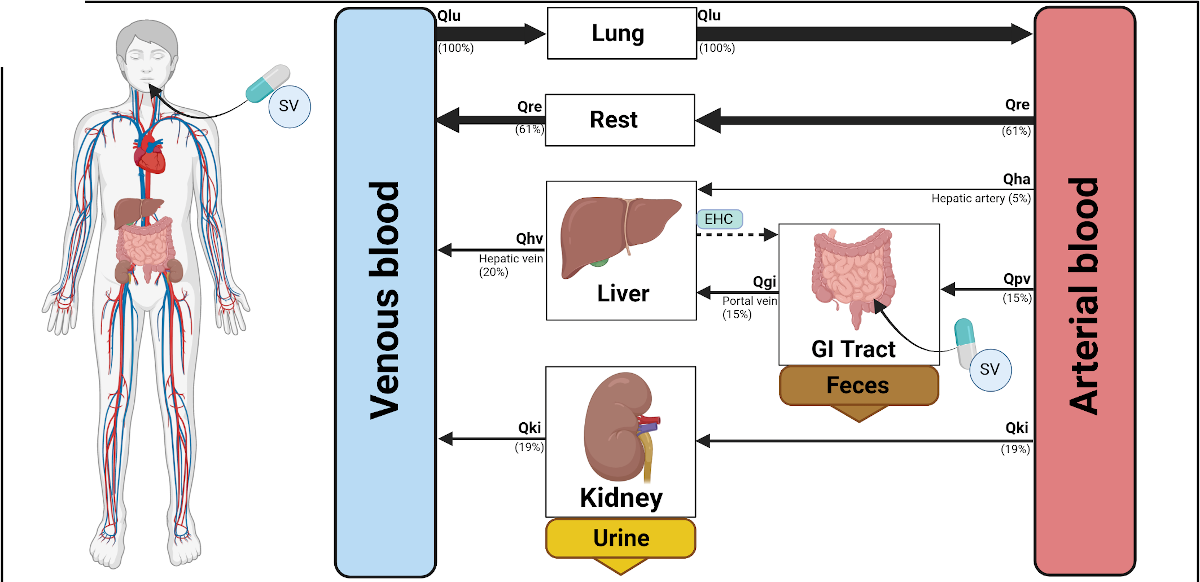
A physiologically-based pharmacokinetic (PBPK) model of sorafenib for investigating the effect of sorafenib parameters in hepatorenal impairment
Internship Report Frances Okibedi (supervisor: Matthias König)
Internship report, July 2024
Liver cancer ranks second in cancer-related deaths thereby making timely diagnosis and personalized treatment crucial. Hepatocellular carcinoma (HCC) patients often get diagnosed at advanced stages, hindering curative options like resection and ablation. Systemic treatment has become the standard therapy and sorafenib has been the primary systemic treatment for advanced HCC for the last ten years, being the first approved drug for this purpose. Sorafenib (Nexavar), is a multi-kinase inhibitor medication approved for primarily treating advanced renal cell carcinoma, hepatocellular carcinoma or other cancers. After oral administration, sorafenib enters hepatocytes via OATP1B-type carriers. It undergoes CYP3A4-mediated oxidative metabolism to sorafenib-N-oxide (M2) and UGT1A9-mediated glucuronidation forming sorafenib-glucuronide (SG). SG is secreted into bile mainly by ABCC2, with a fraction reuptaken by ABCC3, preventing ABCC2 saturation. In bile, SG can be excreted via the feces or converted back to sorafenib by a bacterial β-glucuronidase. Sorafenib is absorbed in the intestine, reentering the systemic circulation, contributing to elimination and detoxification. Sorafenib is excreted mainly by the feces (77%) and urine (19%). Hepatic and renal impairment could have a large impact on the pharmacokinetics of sorafenib. Within this work the pharmacokinetics of sorafenib were analyzed by developing a physiologically-based pharmacokinetic (PBPK) model based on extensive data curation of sorafenib data. The model allowed to simulate the time-concentration courses of sorafenib in various tissues and to calculate the pharmacokinetic parameters for sorafenib under single and multiple dosing regimes. The model was applied to study the dose-dependency of sorafenib treatment and to investigate the effect of hepatic and renal impairment on sorafenib pharmacokinetics.
Keywords:
sorafenib,
PBPK,
pharmacokinetics,
HCC,
liver function,
renal function,
|
|
| 2023 |

|
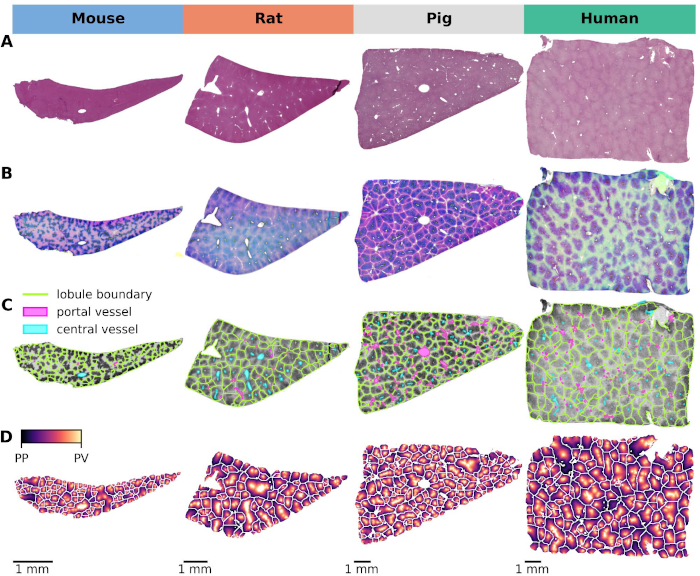
Cross-Species Variability in Lobular Geometry and Cytochrome P450 Hepatic Zonation: Insights into CYP1A2, CYP2E1, CYP2D6 and CYP3A4
Mohamed Albadry, Jonas Kuettner, Jan Grzegorzewski, Olaf Dirsch, Eva Kindler, Robert Klopfleisch, Vaclav Liska, Vladimira Moulisova, Sandra Nickel, Richard Palek, Jachym Rosendorf, Sylvia Saalfeld, Utz Settmacher, Hans-Michael Tautenhahn, Matthias König*, Uta Dahmen* (* equal contribution)
bioRxiv 2023.12.28.573567 (preprint). doi:10.1101/2023.12.28.573567
This study explores the critical interplay between lobular geometry and the zonated distribution of cytochrome P450 (CYP) enzymes across species. We present an innovative approach to assess lobular geometry and zonation patterns using whole slide imaging (WSI). This method allows a detailed, systematic comparison of lobular structures and spatial distribution of key CYP450 enzymes and glutamine synthetase in four different species (mouse, rat, pig, and human). Our results shed light on species differences in lobular geometry and enzymatic zonation, providing critical insights for drug metabolism research. Based on our approach we could determine the minimum number of lobules required for a statistically representative analysis, an important piece of information when evaluating liver biopsies and deriving information from WSI.
Keywords:
liver,
zonation,
cytochrome P450,
lobule geometry,
drug metabolism,
species,
|
|
| 2023 |

|
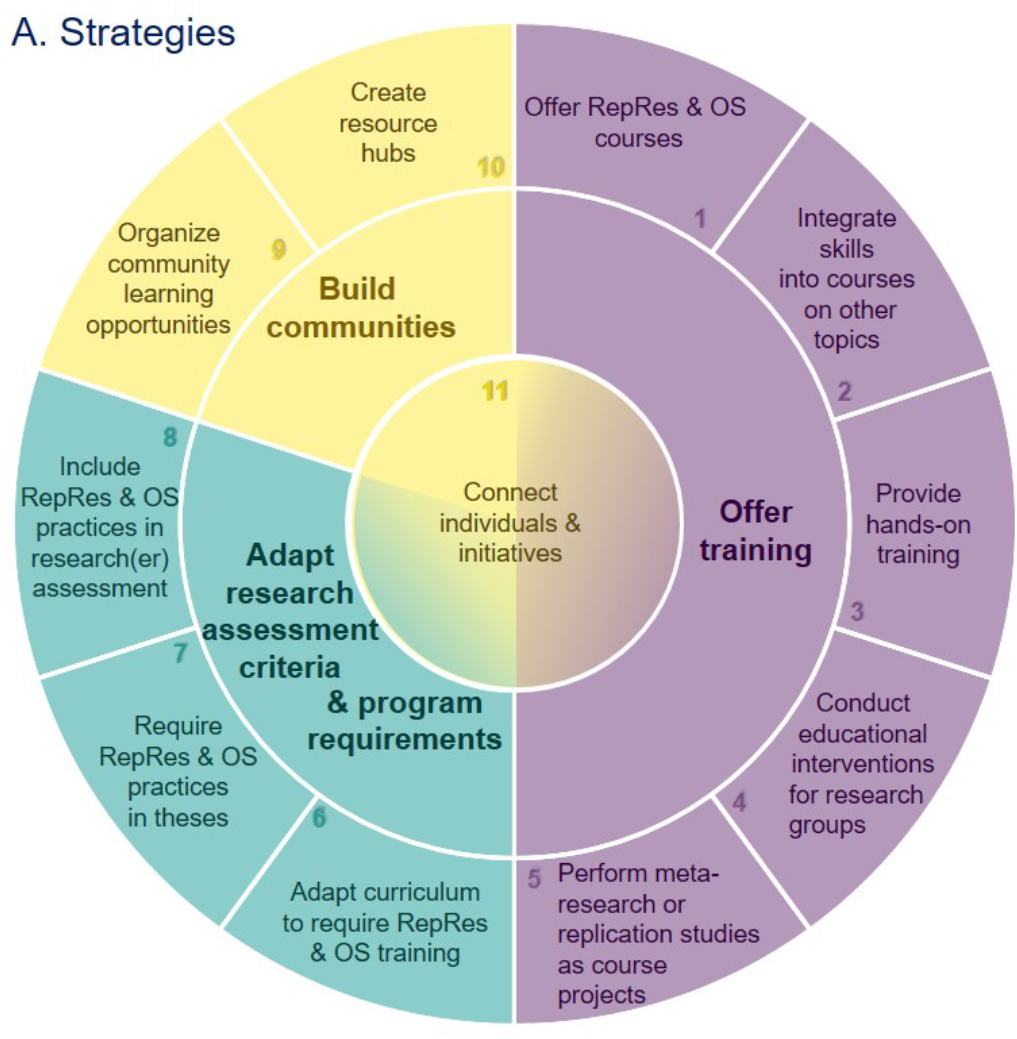
Eleven Strategies for Making Reproducible Research and Open Science Training the Norm at Research Institutions
Friederike E Kohrs, Susann Auer, Alexandra Bannach-Brown, Susann Fiedler, Tamarinde Laura Haven, Verena Heise, Constance Holman, Flavio Azevedo, René Bernard, Armin Bleier, Nicole Bössel, Brian Patrick Cahill, Leyla Jael Castro, Adrian Ehrenhofer, Kristina Eichel, Maximillian Frank, Claudia Frick, Malte Friese, Anne Gärtner, Kerstin Gierend, David Joachim Grüning, Lena Hahn, Maren Hülsemann, Malika Ihle, Sabrina Illius, Laura König, Matthias König, Louisa Kulke, Anton Kutlin, Fritjof Lammers, David MA Mehler, Christoph Miehl, Anett Müller-Alcazar, Claudia Neuendorf, Helen Niemeyer, Florian Pargent, Aaron Peikert, Christina U Pfeuffer, Robert Reinecke, Jan Philipp Röer, Jessica L Rohmann, Alfredo Sánchez-Tójar, Stefan Scherbaum, Elena Sixtus, Lisa Spitzer, Vera Maren Straßburger, Marcel Weber, Clarissa J Whitmire, Josephine Zerna, Dilara Zorbek, Philipp Zumstein, Tracey L Weissgerber
eLife (2023) 12:e89736.. doi:10.7554/eLife.89736
Reproducible research and open science practices have the potential to accelerate scientific progress by allowing others to reuse research outputs, and by promoting rigorous research that is more likely to yield trustworthy results. However, these practices are uncommon in many fields, so there is a clear need for training that helps and encourages researchers to integrate reproducible research and open science practices into their daily work. Here, we outline eleven strategies for making training in these practices the norm at research institutions. The strategies, which emerged from a virtual brainstorming event organized in collaboration with the German Reproducibility Network, are concentrated in three areas: (i) adapting research assessment criteria and program requirements; (ii) training; (iii) building communities. We provide a brief overview of each strategy, offer tips for implementation, and provide links to resources. We also highlight the importance of allocating resources and monitoring impact. Our goal is to encourage researchers – in their roles as scientists, supervisors, mentors, instructors, and members of curriculum, hiring or evaluation committees – to think creatively about the many ways they can promote reproducible research and open science practices in their institutions.
Keywords:
reproducible research,
open science,
training,
|
|
| 2023 |


|
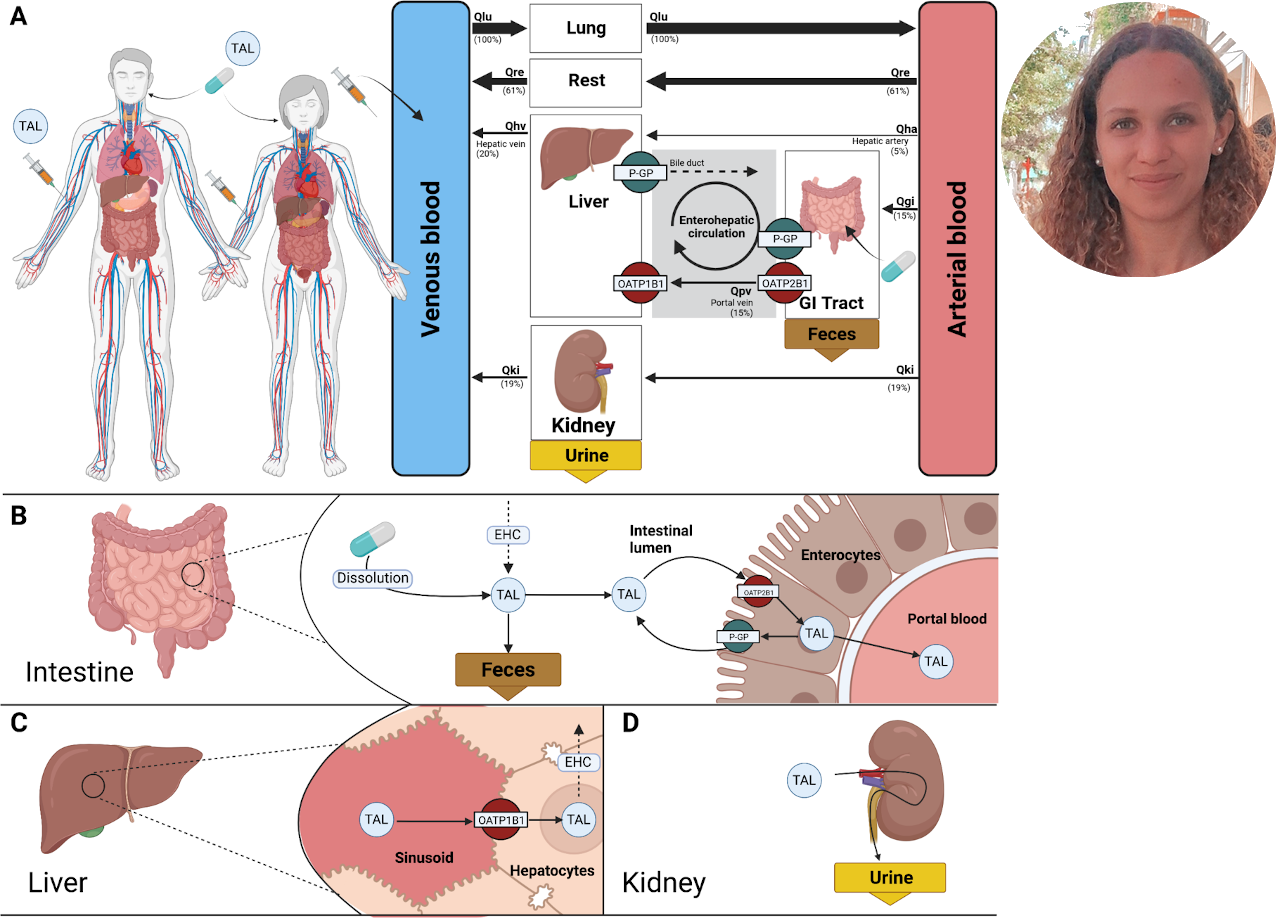
Insights into Intestinal P-glycoprotein Function using Talinolol: A PBPK Modeling Approach
Beatrice Stemmer Mallol, Jan Grzegorzewski, Matthias König
bioRxiv 2023.11.21.568168 (preprint). doi:10.1101/2023.11.21.568168
Talinolol is a cardioselective beta-blocker used in the treatment of various cardiovascular diseases and tachyarrhythmias. Its intestinal absorption is determined by uptake by the organic anion transporting polypeptide 2B1 (OATP2B1) and efflux via P-glycoprotein (P-gp). Talinolol can be taken up via OATP1B1 in the liver, where it enters the enterohepatic circulation. Talinolol is excreted unchanged in the urine and feces. Talinolol is widely used as a probe drug for the intestinal efflux transporter P-gp, which plays a critical role in protecting against potentially toxic substances and facilitating the elimination of xenobiotics. In this work, an extensive database of talinolol pharmacokinetics was established and used to develop and validate a physiologically based pharmacokinetic (PBPK) model of talinolol for P-gp phenotyping. The model was used to investigate the influence of several factors on talinolol pharmacokinetics: (i) inhibition of P-gp via drug-drug interaction; (i) genetic polymorphisms of P-gp; (iii) activity of OATP2B1 and OATP1B1; (iv) effect of disease, namely hepatic and renal impairment; and (v) site-specific distribution of P-gp and OATP2B1 in the intestine. The model accurately predicts the concentration-time profile of talinolol after oral and intravenous administration of single and multiple dosing. Furthermore, the model accurately describes the effect of genetic variants of P-gp on the pharmacokinetics of talinolol, the effect of inhibition of P-gp, the effect of renal impairment, as well as site-specific infusion of talinolol in the intestine. The detailed description of the intestinal absorption of talinolol and the predictions of talinolol pharmacokinetics as a function of hepatorenal impairment provide valuable clinical insights for metabolic phenotyping with talinolol. Both the model and the database are freely available for reuse.
Keywords:
PBPK,
talinolol,
P-glycoprotein,
pharmacokinetics,
OATP2B1,
OATB1B1,
renal impairment,
cirrhosis,
|
|
| 2023 |


|
A pathway model of glucose-stimulated insulin secretion in the pancreatic β-cell
Maheshvare MD., Raha S., König M.*, and Pal D.* (* equal contribution)
Front. Endocrinol. 14:1185656. doi:10.3389/fendo.2023.1185656
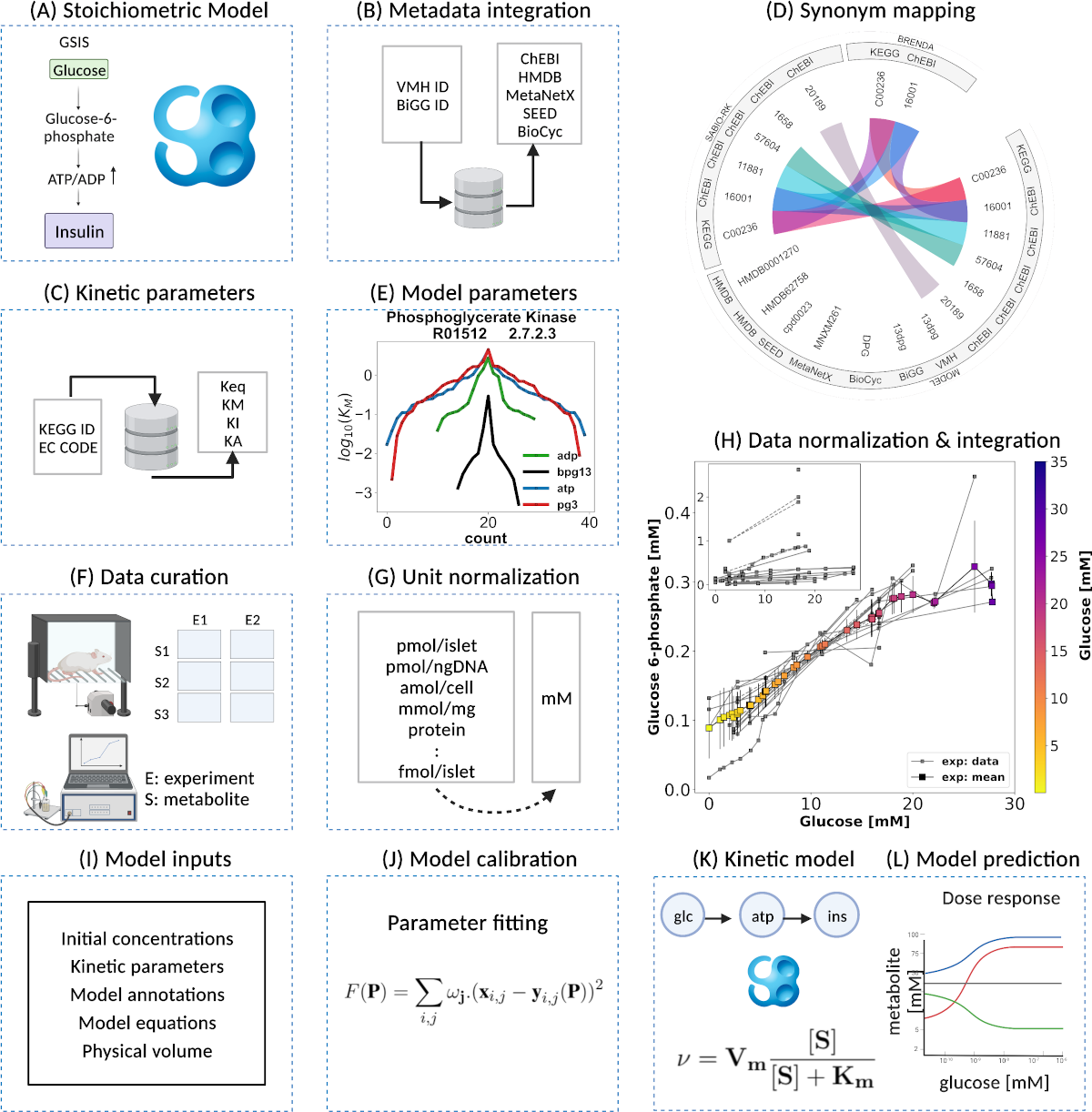
The pancreas plays a critical role in maintaining glucose homeostasis through the secretion of hormones from the islets of Langerhans. Glucose-stimulated insulin secretion (GSIS) by the pancreatic β-cell is the main mechanism for reducing elevated plasma glucose. Here we present a systematic modeling workflow for the development of kinetic pathway models using the Systems Biology Markup Language (SBML). Steps include retrieval of information from databases, curation of experimental and clinical data for model calibration and validation, integration of heterogeneous data including absolute and relative measurements, unit normalization, data normalization, and model annotation. An important factor was the reproducibility and exchangeability of the model, which allowed the use of various existing tools. The workflow was applied to construct a novel data-driven kinetic model of GSIS in the pancreatic β-cell based on experimental and clinical data from 39 studies spanning 50 years of pancreatic, islet, and β-cell research in humans, rats, mice, and cell lines. The model consists of detailed glycolysis and phenomenological equations for insulin secretion coupled to cellular energy state, ATP dynamics and (ATP/ADP ratio). Key findings of our work are that in GSIS there is a glucose-dependent increase in almost all intermediates of glycolysis. This increase in glycolytic metabolites is accompanied by an increase in energy metabolites, especially ATP and NADH. One of the few decreasing metabolites is ADP, which, in combination with the increase in ATP, results in a large increase in ATP/ADP ratios in the β-cell with increasing glucose. Insulin secretion is dependent on ATP/ADP, resulting in glucose-stimulated insulin secretion. The observed glucose-dependent increase in glycolytic intermediates and the resulting change in ATP/ADP ratios and insulin secretion is a robust phenomenon observed across data sets, experimental systems and species. Model predictions of the glucose-dependent response of glycolytic intermediates and biphasic insulin secretion are in good agreement with experimental measurements. Our model predicts that factors affecting ATP consumption, ATP formation, hexokinase, phosphofructokinase, and ATP/ADP-dependent insulin secretion have a major effect on GSIS. In conclusion, we have developed and applied a systematic modeling workflow for pathway models that allowed us to gain insight into key mechanisms in GSIS in the pancreatic β-cell.
Keywords:
glucose-stimulated insulin secretion,
GSIS,
glycolysis,
pancreas,
kinetic model,
systems biology,
|
|
| 2023 |


|
A physiologically based pharmacokinetic (PBPK) model of the probe drug talinolol for the characterization of intestinal P-glycoprotein
Bachelor Thesis Beatrice Stemmer Mallol (supervisor: Matthias König)
Bachelor Thesis, July 2023
Talinolol is a cardioselective beta-blocker used for the treatment of various cardiovascular diseases and tachyarrhythmia. The gastrointestinal absorption of talinolol is determined via its uptake in the intestine via the organic anion transporting polypeptide 2B1 (OATP2B1) and its efflux via the P-glycoprotein (P-gp). After intestinal absorption talinolol can be transported into the liver via OATP1B1 talinolol where it undergoes enterohepatic circulation. Talinolol is excreted unchanged in the urine and feces. In addition to its clinical application, talinolol is widely used as a probe drug for the intestinal efflux transporter P-glycoprotein. P-gp plays a crucial role in the human body as it is expressed in various tissues to protect against potentially toxic substances, facilitating the elimination of xenobiotics. The application of talinolol for P-gp phenotyping enables evaluation of factors influencing P-gp-mediated transport in vivo such as genetic polymorphisms of P-gp as well as the distribution of P-gp along the intestine. Within this thesis, an extensive dataset of talinolol pharmacokinetics was established and used to develop a physiologically based pharmacokinetic (PBPK) model for talinolol. The model was applied to investigate the influence of various factors on the pharmacokinetics of talinolol, including: (i) genetic variants of P-gp; (ii) enzymatic activity of the transporters OATP2B1 and OATP1B1; (iii) site-specific distribution of P-gp and OATP2B1 proteins in the intestine, and (iv) the impact of diseases such as liver cirrhosis and renal dysfunction. The model enables precise predictions of the concentration-time profile of talinolol in various tissues following oral or intravenous administration. Furthermore, the model accurately describes the effect of genetic variants of P-gp on the pharmacokinetics of talinolol. The detailed description of the limiting intestinal absorption of intestinal provided by the model, along with the precise prediction of talinolol`s pharmacokinetics in different renal functions, holds significant clinical relevance.
Keywords:
talinolol,
PBPK,
pharmacokinetics,
OATP2B1,
OATP1B1,
P-glycoprotein,
liver function,
renal function,
|
|
| 2023 |


|
A physiologically based pharmacokinetic model for CYP2E1 phenotyping via chlorzoxazone
J. Küttner, J. Grzegorzewski, HM. Tautenhahn, M. König
bioRxiv 2023.04.12.536571 (preprint). doi:10.1101/2023.04.12.536571
The cytochrome P450 (CYP) superfamily of enzymes plays a critical role in the metabolism of drugs, toxins, and endogenous and exogenous compounds. The activity of CYP enzymes can be influenced by a variety of factors, including genetics, diet, age, environmental factors, and disease. Among the major isoforms, CYP2E1 is of particular interest due to its involvement in the metabolism of various low molecular weight chemicals, including alcohols, pharmaceuticals, industrial solvents, and halogenated anesthetics. Metabolic phenotyping of CYPs based on the elimination of test compounds is a useful method for assessing in vivo activity, with chlorzoxazone being the primary probe drug for phenotyping of CYP2E1. The aim of this work was to investigate the effect of changes in CYP2E1 level and activity, ethanol consumption, ethanol abstinence, and liver impairment on the results of metabolic phenotyping with chlorzoxazone. To accomplish this, an extensive pharmacokinetic dataset for chlorzoxazone was established and a physiologically based pharmacokinetic (PBPK) model of chlorzoxazone and its metabolites, 6-hydroxychlorzoxazone and chlorzoxazone-O-glucuronide, was developed and validated. The model incorporates the effect of ethanol consumption on CYP2E1 levels and activity by extending the model with a core ethanol pharmacokinetic model and a CYP2E1 turnover model. The model accurately predicts pharmacokinetic data from several clinical studies and is able to estimate the effect of changes in CYP2E1 levels and activity on chlorzoxazone pharmacokinetics. Regular ethanol consumption induces CYP2E1 over two to three weeks, resulting in increased conversion of chlorzoxazone to 6-hydroxychlorzoxazone and a higher 6-hydroxychlorzoxazone/chlorzoxazone metabolic ratio. After ethanol withdrawal, CYP2E1 levels return to baseline within one week. Importantly, liver impairment has an opposite effect, resulting in reduced liver function via CYP2E1. In alcoholics with liver impairment who also consume ethanol, these factors will have opposite confounding effects on metabolic phenotyping with chlorzoxazone.
Keywords:
PBPK,
chlorzoxazone,
CYP2E1,
alcohol,
ethanol,
pharmacokinetics,
|
|
| 2023 |


|
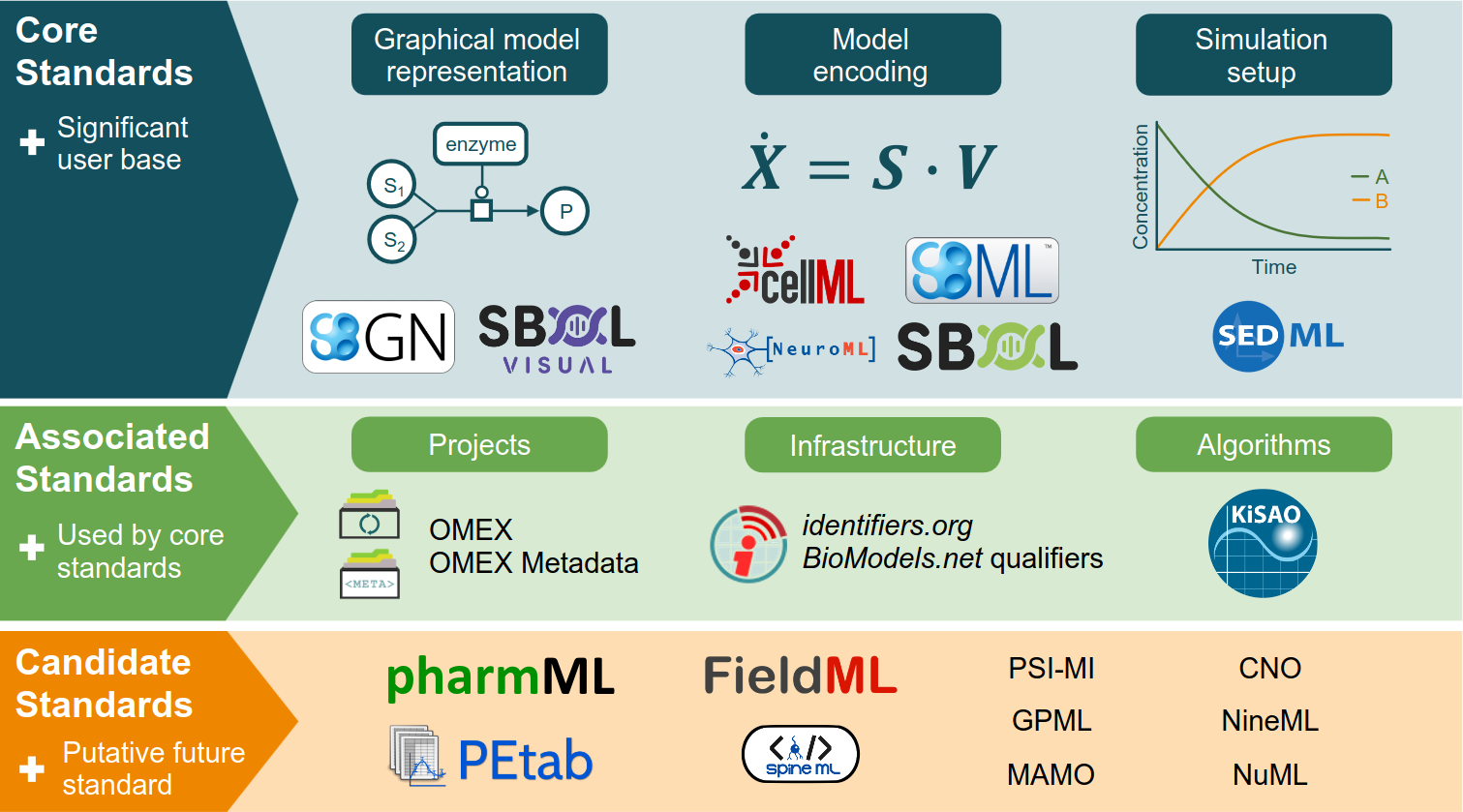
Specifications of Standards in Systems and Synthetic Biology: Status and Developments in 2022 and the COMBINE meeting 2022
M. König, P. Gleeson, M. Golebiewski, T. Gorochowski, M. Hucka, S. Keating, C. Myers, D. Nickerson, F. Schreiber
J Integr Bioinform. 2023 Mar 29;20(1).. doi:10.1515/jib-2023-0004. pmid:36989443
This special issue of the Journal of Integrative Bioinformatics contains updated specifications of COMBINE standards in systems and synthetic biology. The 2022 special issue presents three updates to the standards: CellML 2.0.1, SBML Level 3 Package: Spatial Processes, Version 1, Release 1, and Synthetic Biology Open Language (SBOL) Version 3.1.0. This document can also be used to identify the latest specifications for all COMBINE standards. In addition, this editorial provides a brief overview of the COMBINE 2022 meeting in Berlin.
Keywords:
COMBINE,
standardization,
SBML,
SEDML,
OMEX,
|
|
| 2023 |


|
standard-GEM: standardization of open-source genome-scale metabolic models
Mihail Anton, Eivind Almaas, Rui Benfeitas, Sara Benito-Vaquerizo, Lars M. Blank, Andreas Dräger, John M. Hancock, Cheewin Kittikunapong, Matthias König, Feiran Li, Ulf W. Liebal, Hongzhong Lu, Hongwu Ma, Radhakrishnan Mahadevan, Adil Mardinoglu, Jens Nielsen, Juan Nogales, Marco Pagni, Jason A. Papin, Kiran Raosaheb Patil, Nathan D. Price, Jonathan L. Robinson, Benjamín J. Sánchez, Maria Suarez-Diez, Snorre Sulheim, L. Thomas Svensson, Bas Teusink, Wanwipa Vongsangnak, Hao Wang, Ahmad A. Zeidan, Eduard J. Kerkhoven
bioRxiv 2023.03.21.512712 (preprint). doi:10.1101/2023.03.21.512712
The field of metabolic modelling at the genome-scale continues to grow with more models being created and curated. This comes with an increasing demand for adopting common principles regarding transparency and versioning, in addition to standardisation efforts regarding file formats, annotation and testing. Here, we present a standardised template for git-based and GitHub-hosted genome-scale metabolic models (GEMs) supporting both new models and curated ones, following FAIR principles (findability, accessibility, interoperability, and reusability), and incorporating best-practices. standard-GEM facilitates the reuse of GEMs across web services and platforms in the metabolic modelling field and enables automatic validation of GEMs. The use of this template for new models, and its adoption for existing ones, paves the way for increasing model quality, openness, and accessibility with minimal effort.
Keywords:
genome-scale metabolic model,
open source,
versioning,
|
|
| 2023 |


|
Simvastatin therapy in different subtypes of hypercholesterolemia - a physiologically based modelling approach
F. Bartsch, J. Grzegorzewski, H. Pujol, HM. Tautenhahn, M. König
medRxiv 2023.02.01.23285358 (preprint). doi:10.1101/2023.02.01.23285358
Hypercholesterolemia is a multifaceted plasma lipid disorder with heterogeneous causes including lifestyle and genetic factors. A key feature of hypercholesterolemia is elevated plasma levels of low-density lipoprotein cholesterol (LDL-C). Several genetic variants have been reported to be associated with hypercholesterolemia, known as familial hypercholesterolemia (FH). Important variants affect the LDL receptor (LDLR), which mediates the uptake of LDL-C from the plasma, apoliporotein B (APOB), which is involved in the binding of LDL-C to the LDLR, and proprotein convertase subtilisin/kexin type 9 (PCSK9), which modulates the degradation of the LDLR. A typical treatment for hypercholesterolemia is statin medication, with simvastatin being one of the most commonly prescribed statins. In this work, the LDL-C lowering therapy with simvastatin in hypercholesterolemia was investigated using a computational modeling approach. A physiologically based pharmacokinetic model of simvastatin integrated with a pharmacodynamic model of plasma LDL-C (PBPK/PD) was developed based on extensive data curation. A key component of the model is LDL-C turnover by the liver, consisting of: hepatic cholesterol synthesis with the key enzymes HMG-CoA reductase and HMG-CoA synthase; cholesterol export from the liver as VLDL-C; de novo synthesis of LDLR; transport of LDLR to the membrane; binding of LDL-C by LDLR via APOB; endocytosis of the LDLR-LDL-C complex; recycling of LDLR from the complex. The model was applied to study the effects of simvastatin therapy in hypercholesterolemia due to different causes in the LDLR pathway corresponding to different subtypes of hypercholesterolemia. Model predictions of LDL-C lowering therapy were validated with independent clinical data sets. Key findings are: (i) hepatic LDLR turnover is highly heterogeneous among FH classes; (ii) despite this heterogeneity, simvastatin therapy results in a consistent reduction in plasma LDL-C regardless of class; and (iii) simvastatin therapy shows a dose-dependent reduction in LDL-C. Our model suggests that the underlying cause of hypercholesterolemia does not influence simvastatin therapy. Furthermore, our model supports the treatment strategy of stepwise dose adjustment to achieve target LDL-C levels. Both the model and the database are freely available for reuse.
Keywords:
simvastatin,
hypercholesterolemia,
LDL-cholesterol,
LDL-receptor,
physiologically based pharmacokinetic model,
pharmacokinetics,
pharmacodynamics,
PK/PD,
|
|
| 2023 |


|
Physiologically based pharmacokinetic (PBPK) modeling for dynamical liver function tests and CYP phenotyping
Jan Grzegorzewski (supervisor: Matthias König)
PhD Thesis, Jan 2023 (submitted)
Cytochrome P450 (CYP) phenotyping and dynamic liver function testing are essential methods in clinical practice. These methods utilize the pharmacokinetics (PK) of test substances and their metabolites to gain insight into the liver’s metabolic capacity and the activity of enzymes and transporters. Despite an extensive body of literature, many aspects affecting liver function and CYP activity are not well understood. Liver function tests are not only influenced by numerous characteristics of a studied subject but also by the specifics of individual study procedures. A key challenge is to disentangle the various factors which influence the outcome of the measurements from each other to study their influence on the dynamic liver function and CYP phenotype. In this work, the challenge was addressed through meta-analysis and physiologically based pharmacokinetic (PBPK) modeling. As a foundation, an open pharmacokinetics database (https://pk-db.com) was developed and pharmacokinetics data were curated for a wide range of test substances. To my knowledge, PK-DB currently contains the largest open pharmacokinetic dataset on substances used for phenotyping and dynamical liver function testing. The dataset allowed for identifying and quantifying demographic and racial bias (sex, ethnicity, age, health), reporting errors, and inconsistencies in pharmacokinetic literature. Based on the data, a caffeine pharmacokinetics meta-analysis was conducted concerning various factors affecting liver function and CYP1A2 activity. In particular, meta-analysis and data integration solidified existing knowledge on the effects of smoking, oral contraceptives, multiple diseases, and co-medications on caffeine pharmacokinetics. Similarly, the measurement accuracy of caffeine concentration in saliva versus plasma was quantified, and the effect of dosing amount and sampling timing for phenotyping were analyzed. In addition, the impact of CYP2D6 polymorphism was investigated. Therefore, a PBPK model of dextromethorphan (DXM) and its metabolites dextrorphan (DXO) and dextrorphan O-glucuronide (DXO-Glu) was developed, and calibrated and validated with pharmacokinetics data. The variability in CYP activity was modeled based on in vitro data. The model can predict individual plasma concentrations and urinary amounts of DXM, DXO, and DXO-Glu and the metabolic phenotype based on the individual’s CYP2D6 genotype and physiological characteristics. The analyses suggest that most of the variability in the pharmacokinetics of dextromethorphan can be attributed to the variability in CYP2D6 and CYP3A4 enzyme kinetics. Among various other investigations, the influence of ethnicity on CYP2D6 activity was also investigated. Contributions to PK data curation and PBPK model development were also made for other phenotyping and liver function test substances (chlorzoxazone, codeine, diazepam, galactose, indocyanine green (ICG), metoprolol, midazolam, omeprazole, pravastatin, simvastatin, talinolol, and torasemide). For ICG, in particular, the impact of hepatic blood flow, cardiac output, and body weight, as well as the survival probability after partial hepatectomy, were investigated by PBPK modeling. Notably, the studying of the various test substances was only made possible by a systematic and standardized workflow that facilitated data integration, data sharing, the creation of reproducible PBPK models, and the standardized integration of data and models. In conclusion, a pharmacokinetic database, methods, and workflows for the analysis of test compounds used in dynamical liver function testing and CYP phenotyping were established. Factors affecting CYP phenotyping and liver function testing were investigated by meta-analysis and PBPK modeling. The models developed in this effort have the potential to impact personalized medicine and to increase the precision of dynamic liver function tests in clinics.
Keywords:
liver function,
metabolic phenotyping,
pharmacokinetics,
PBPK,
|
|
| 2023 |



|
libRoadRunner 2.0: A High-Performance SBML Simulation and Analysis Library
Ciaran Welsh, Jin Xu, Lucian Smith, Matthias König, Kiri Choi, Herbert M. Sauro
Bioinformatics. 2023 Jan 1;39(1):btac770. doi:10.1093/bioinformatics/btac770. pmid:36478036
Motivation: This paper presents libRoadRunner 2.0, an extensible, high-performance, cross-platform, open-source software library for the simulation and analysis of models expressed using the Systems Biology Markup Language (SBML). Results: libRoadRunner is a self-contained library, able to run either as a component inside other tools via its C ++, C and Python APIs, or interactively through its Python or Julia interface. libRoadRunner uses a custom Just-In-Time (JIT) compiler built on the widely-used LLVM JIT compiler framework. It compiles SBML-specified models directly into native machine code for a large variety of processors, making it fast enough to simulate extremely large models or repeated runs in reasonable timeframes. libRoadRunner is flexible, supporting the bulk of the SBML specification (except for delay and nonlinear algebraic equations) as well as several SBML extensions such as hierarchical composition and probability distributions. It offers multiple deterministic and stochastic integrators, as well as tools for steady-state, sensitivity, stability and structural analyses. Availability: libRoadRunner binary distributions for Windows, Mac OS, and Linux, Julia and Python bindings, source code, and documentation are all available at https://github.com/sys-bio/roadrunner, and Python bindings are also available via pip. The source code can be compiled for the supported systems as well as in principle any system supported by LLVM-13, such as ARM-based computers like the Raspberry Pi. The library is licensed under the Apache License Version 2.0.
Keywords:
computer simulation,
software,
systems biology,
|
|
| 2022 |

|
Cytochrome p450 enzymes in periportal steatosis: Modulation of drug metabolizing activity but not of pericentral expression pattern
M. Albadry, S. Höpfl, N. Ehteshamzad, M. König, M. Böttcher, J. Neumann, A. Lupp, O. Dirsch, N. Radde, B. Christ, M. Christ, L.O. Schwen, H. Laue, R. Klopfleisch and Uta Dahmen
Sci Rep. 2022 Dec 17;12(1):21825. doi:10.1038/s41598-022-26483-6. pmid:36528753
Little is known about the impact of morphological disorders in distinct zones on metabolic zonation. It was described recently that periportal fibrosis did affect the expression of CYP proteins, a set of pericentrally located drug-metabolizing enzymes. Here, we investigated whether periportal steatosis might have a similar effect. Periportal steatosis was induced in C57BL6/J mice by feeding a high-fat diet with low methionine/choline content for either two or four weeks. Steatosis severity was quantified using image analysis. Triglycerides and CYP activity were quantified in photometric or fluorometric assay. The dis- tribution of CYP3A4, CYP1A2, CYP2D6, and CYP2E1 was visualized by immunohistochemistry. Pharmacokinetic parameters of test drugs were determined after injecting a drug cocktail (caffeine, codeine, and midazolam). The dietary model resulted in moderate to severe mixed steatosis confined to periportal and midzonal areas. Periportal steatosis did not affect the zonal distribution of CYP expression but the activity of selected CYPs was associated with steatosis severity. Caffeine elimination was accelerated by microvesicular steatosis, whereas midazolam elimination was delayed in macrovesicular steatosis. In summary, periportal steatosis affected parameters of pericentrally located drug metabolism. This observation calls for fur- ther investigations of the highly complex interrelationship between steatosis and drug metabolism and underlying signaling mechanisms.
Keywords:
cytochrome P-450 Enzyme System,
fatty Liver,
mice,
pharmacokinetics,
|
|
| 2022 |


|
Physiologically based pharmacokinetic (PBPK) modeling of the role of CYP2D6 polymorphism for metabolic phenotyping with dextromethorphan
Grzegorzewski, J., Brandhorst, J., König, M.
Front Pharmacol. 2022 Oct 24;13:1029073. doi:10.3389/fphar.2022.1029073. pmid:36353484
The cytochrome P450 2D6 (CYP2D6) is a key xenobiotic-metabolizing enzyme involved in the clearance of many drugs. Genetic polymorphisms in CYP2D6 contribute to the large inter-individual variability in drug metabolism and could affect metabolic phenotyping of CYP2D6 probe substances such as dextromethorphan (DXM). To study this question, we (i) established an extensive pharmacokinetics dataset for DXM; and (ii) developed and validated a physiologically based pharmacokinetic (PBPK) model of dextromethorphan (DXM) and its metabolites dextrorphan (DXO) and dextrorphan O-glucuronide (DXO-Glu) based on the data. Drug-gene interactions (DGI) were introduced by accounting for changes in CYP2D6 enzyme kinetics depending on activity score (AS), which in combination with AS for individual polymorphisms allowed us to model of CYP2D6 gene variants. Variability in CYP3A4 and CYP2D6 activity was modeled based on in vitro data from human liver microsomes. Model predictions are in very good agreement with pharmacokinetics data for CYP2D6 polymorphisms, CYP2D6 activity as measured by AS, and CYP2D6 metabolic phenotypes (UM, EM, IM, PM). The model was applied to investigate the genotype-phenotype association and the role of CYP2D6 polymorphisms for metabolic phenotyping with dextromethorphan using the urinary cumulative metabolic ratio DXM/(DXO+DXO-Glu) (UCMR). The effect of parameters on UCMR was studied via sensitivity analysis. Model predictions indicate very good robustness against the intervention protocol (i.e. application form, dosing amount, dissolution rate, and sampling time) and good robustness against physiological variation. The model is capable of estimating the UCMR dispersion within and across populations depending on activity scores. Moreover, the distribution of UCMR and the risk of genotype-phenotype mismatch could be estimated for populations with known CYP2D6 genotype frequencies. The model can be applied as a tool for individual prediction of UCMR and metabolic phenotype based on CYP2D6 genotype. Both, model and database are freely available for reuse.
Keywords:
Dextromethorphan,
CYP2D6,
Physiologically based pharmacokinetic model,
PBPK,
Pharmacokinetics,
Pharmacogenomics,
|
|
| 2022 |



|
FAIR Sharing of Reproducible Models of Epidemic and Pandemic Forecast
Ramachandran, K.*; König, M.*; Scharm, M.; Nguyen, T.V.N.; Hermjakob, H.; Waltemath, D.; Malik Sheriff, R.S. (* equal contribution)
Preprints 2022, 2022060137 (preprint). doi:10.20944/preprints202206.0137.v1
A major challenge for the dissemination, replication, and reuse of epidemiological forecasting studies during COVID-19 pandemics is the lack of clear guidelines and platforms to exchange models in a Findable, Accessible, Interoperable, and Reusable (FAIR) manner, facilitating reproducibility of research outcomes. During the beginning of pandemics, models were developed in diverse tools that were not interoperable, opaque without traceability and semantics, and scattered across various platforms - making them hard to locate, infer and reuse. In this work, we demonstrate that implementing the standards developed by the systems biology community to encode and share COVID-19 epidemiological models can serve as a roadmap to implement models as a tool in medical informatics, in general. As a proof-of-concept, we encoded and shared 24 epidemiological models using the standard format for model exchange in systems biology, annotated them with cross-references to data resources, packed up all associated files in COMBINE archives for easy sharing, and finally, disseminated the models through BioModels repository to significantly enhance their reproducibility and repurposing potential. We recommend the use of systems biology standards to encode and share models of epidemic and pandemic forecasts to improve their findability, accessibility, interoperability, reusability, and reproducibility.
Keywords:
FAIR,
epidemiology,
models,
pandemic forecast,
SIR modelling,
standards,
|
|
| 2022 |



|
BioSimulators: a central registry of simulation engines and services for recommending specific tools
Shaikh B, Smith LP, Vasilescu D, Marupilla G, Wilson M, Agmon E, Agnew H, Andrews SS, Anwar A, Beber ME, Bergmann FT, Brooks D, Brusch L, Calzone L, Choi K, Cooper J, Detloff J, Drawert B, Dumontier M, Ermentrout GB, Faeder JR, Freiburger AP, Fröhlich F, Funahashi A, Garny A, Gennari JH, Gleeson P, Goelzer A, Haiman Z, Hasenauer J, Hellerstein JL, Hermjakob H, Hoops S, Ison JC, Jahn D, Jakubowski HV, Jordan R, Kalaš M, König M, Liebermeister W, Sheriff RSM, Mandal S, McDougal R, Medley JK, Mendes P, Müller R, Myers CJ, Naldi A, Nguyen TVN, Nickerson DP, Olivier BG, Patoliya D, Paulevé L, Petzold LR, Priya A, Rampadarath AK, Rohwer JM, Saglam AS, Singh D, Sinha A, Snoep J, Sorby H, Spangler R, Starruß J, Thomas PJ, van Niekerk D, Weindl D, Zhang F, Zhukova A, Goldberg AP, Schaff JC, Blinov ML, Sauro HM, Moraru II, Karr JR.
Nucleic Acids Res. 2022 May 7;50(W1):W108–14. doi:10.1093/nar/gkac331. pmid:35524558
Computational models have great potential to accelerate bioscience, bioengineering, and medicine. However, it remains challenging to reproduce and reuse simulations, in part, because the numerous formats and methods for simulating various subsystems and scales remain siloed by different software tools. For example, each tool must be executed through a distinct interface. To help investigators find and use simulation tools, we developed BioSimulators (https://biosimulators.org), a central registry of the capabilities of simulation tools and consistent Python, command-line and containerized interfaces to each version of each tool. The foundation of BioSimulators is standards, such as CellML, SBML, SED-ML and the COMBINE archive format, and validation tools for simulation projects and simulation tools that ensure these standards are used consistently. To help modelers find tools for particular projects, we have also used the registry to develop recommendation services. We anticipate that BioSimulators will help modelers exchange, reproduce, and combine simulations.
Keywords:
reproducibility,
software,
simulation,
|
|
| 2022 |


|
A physiologically based model of pravastatin - The role of genotypes and hepatic or renal impairment on the pharmacokinetics of pravastatin
Helena Leal Pujol (supervisor: Matthias König)
Bachelor Thesis, May 2022
Hypercholesterolaemia, i.e., elevated plasma levels of cholesterol, is a major risk factor for cardiovascular disease, the leading cause of death globally. Hypercholesterolaemia can be treated using statins, a class of medications which inhibit HMG-CoA reductase, a major enzyme in cholesterol synthesis. Pravastatin is a statin used to reduce total and low-density plasma cholesterol levels and increase high-density plasma cholesterol levels in hypercholesterolaemic patients. Pravastatin is absorbed from the small intestine by the transporter OATP2B1 and subsequently transported in the liver via OATP1B1 from where it can be exported in the bile via the enzymatic exporter MRP2. Pravastatin can be excreted either in the urine via the kidneys or in the faeces due to incomplete absorption. Hepatic and renal impairment could have a large impact on the pharmacokinetics of statins as could have genetic variants of the transporters OATP2B1, OATP1B1 and MRP2. Within this thesis the pharmacokinetics of pravastatin were analysed by developing a physiologically-based pharmacokinetics model based on extensive data curation of pravastatin data. The model allows to simulate the time-concentration courses of pravastatin in various tissues and to calculate the pharmacokinetic parameters for pravastatin. Furthermore, the model was applied to investigate the effects of genotypes of the enzymatic transporters and hepatic and renal impairment on the pharmacokinetics of pravastatin. Thus, key questions such as, how pravastatin therapy would be affected in renal or hepatic disease, as well as how pravastatin therapy should be adapted based on genotypes, find an answer in this work.
Keywords:
pravastatin,
PBPK,
pharmacokinetics,
OATP2B1,
OATP1B1,
MRP2,
liver function,
renal function,
|
|
| 2022 |



|
Dynamic Flux Balance Analysis Models in SBML
Matthias König*, Leandro H. Watanabe*, Jan Grzegorzewski, and Chris J. Myers
* equal contribution
bioRxiv 245076 (preprint, submitted). doi:10.1101/245076
Computational models in systems biology and systems medicine are typically simulated using a single formalism such as ordinary differential equations (ODE). However, more complex models require the coupling of multiple formalisms since different biological phenomena are better described by different methods. For example, metabolism in steady state is often modeled using flux-balance analysis (FBA) whereas dynamic changes of model components are better described via ODEs. The coupling of FBA and ODE modeling formalisms results in dynamic FBA models. A major challenge is how to describe such hybrid models that couple multiple formalisms in a standardized way so that they can be exchanged between tools and simulated consistently in a reproducible manner. This paper presents a scheme for encoding and implementation of dynamic FBA models in the Systems Biology Markup Language (SBML), thereby enabling the exchange of multi-framework computational models between software tools. We demonstrate the feasibility of the approach using various example models and show that different tools are able to simulate the hybrid models and agree on the results. As part of this work, two independent implementations of a multi-framework simulation method for dynamic FBA have been developed supporting such models: iBioSim and sbmlutils.
Keywords:
dynamic flux balance analysis,
DFBA,
flux balance analysis (FBA),
ordinary differential equations (ODE),
static optimization approach (SOA),
|
|
| 2022 |


|
Pharmacokinetics of caffeine: A systematic analysis of reported data for application in metabolic phenotyping and liver function testing
Jan Grzegorzewski, Florian Bartsch, Adrian Köller, and Matthias König
Frontiers in Pharmacology 2022, Vol12. doi:10.3389/fphar.2021.752826. pmid:35280254
Caffeine is by far the most ubiquitous psychostimulant worldwide found in tea, coffee, cocoa, energy drinks, and many other beverages and food. Caffeine is almost exclusively metabolized in the liver by the cytochrome P-450 enzyme system to the main product paraxanthine and the additional products theobromine and theophylline. Besides its stimulating properties, two important applications of caffeine are metabolic phenotyping of cytochrome P450 1A2 (CYP1A2) and liver function testing. An open challenge in this context is to identify underlying causes of the large inter-individual variability in caffeine pharmacokinetics. Data is urgently needed to understand and quantify confounding factors such as lifestyle (e.g., smoking), the effects of drug-caffeine interactions (e.g., medication metabolized via CYP1A2), and the effect of disease. Here we report the first integrative and systematic analysis of data on caffeine pharmacokinetics from 141 publications and provide a comprehensive high-quality data set on the pharmacokinetics of caffeine, caffeine metabolites, and their metabolic ratios in human adults. The data set is enriched by meta-data on the characteristics of studied patient cohorts and subjects (e.g., age, body weight, smoking status, health status), the applied interventions (e.g., dosing, substance, route of application), measured pharmacokinetic time-courses, and pharmacokinetic parameters (e.g., clearance, half-life, area under the curve). We demonstrate via multiple applications how the data set can be used to solidify existing knowledge and gain new insights relevant for metabolic phenotyping and liver function testing based on caffeine. Specifically, we analyzed 1) the alteration of caffeine pharmacokinetics with smoking and use of oral contraceptives; 2) drug-drug interactions with caffeine as possible confounding factors of caffeine pharmacokinetics or source of adverse effects; 3) alteration of caffeine pharmacokinetics in disease; and 4) the applicability of caffeine as a salivary test substance by comparison of plasma and saliva data. In conclusion, our data set and analyses provide important resources which could enable more accurate caffeine-based metabolic phenotyping and liver function testing.
Keywords:
CYP1A2,
caffeine,
drug-disease interactions,
drug-drug interactions,
liver function test,
oral contraceptives,
pharmacokinetics,
smoking,
|
|
| 2021 |


|
Prediction of survival after hepatectomy using a physiologically based pharmacokinetic model of indocyanine green liver function tests
Adrian Köller, Jan Grzegorzewski, Michael Tautenhahn, Matthias König
Front. Physiol., 22 November 2021. doi:10.3389/fphys.2021.730418. pmid:34880771
The evaluation of hepatic function and functional capacity of the liver are essential tasks in hepatology as well as in hepatobiliary surgery. Indocyanine green (ICG) is a widely applied test compound that is used in clinical routine to evaluate hepatic function. Important questions for the functional evaluation with ICG in the context of hepatectomy are how liver disease such as cirrhosis alters ICG elimination, and if postoperative survival can be predicted from preoperative ICG measurements. Within this work a physiologically based pharmacokinetic (PBPK) model of ICG was developed and applied to the prediction of the effects of a liver resection under various degrees of cirrhosis. For the parametrization of the computational model and validation of model predictions a database of ICG pharmacokinetic data was established. The model was applied (i) to study the effect of liver cirrhosis and liver resection on ICG pharmacokinetics; and (ii) to evaluate the model-based prediction of postoperative ICG-R15 (retention ratio 15 min after administration) as a measure for postoperative outcome. Key results are the accurate prediction of changes in ICG pharmacokinetics caused by liver cirrhosis and postoperative changes of ICG-elimination after liver resection, as validated with a wide range of data sets. Based on the PBPK model, individual survival after liver resection could be classified, demonstrating its potential value as a clinical tool.
Keywords:
computational model,
hepatectomy,
indocyanine green,
liver cirrhosis,
liver function,
liver resection,
mathematical model,
pharmacokinetics,
|
|
| 2021 |


|
Physiologically based modeling of the effect of physiological and anthropometric variability on indocyanine green based liver function tests
Adrian Köller, Jan Grzegorzewski and Matthias König
Front Physiol. 2021 Nov 22;12:757293. doi:10.3389/fphys.2021.757293. pmid:34880776
Accurate evaluation of liver function is a central task in hepatology. Dynamic liver function tests (DLFT) based on the time-dependent elimination of a test substance provide an important tool for such a functional assessment. These tests are used in the diagnosis and monitoring of liver disease as well as in the planning of hepatobiliary surgery. A key challenge in the evaluation of liver function with DLFTs is the large inter-individual variability. Indocyanine green (ICG) is a widely applied test compound used for the evaluation of liver function. After an intravenous administration, pharmacokinetic (PK) parameters are calculated from the plasma disappearance curve of ICG which provide an estimate of liver function. The hepatic elimination of ICG is affected by physiological factors such as hepatic blood flow or binding of ICG to plasma proteins, anthropometric factors such as body weight, age, and sex, or the protein amount of the organic anion-transporting polypeptide 1B3 (OATP1B3) mediating the hepatic uptake of ICG. Being able to account for and better understand these various sources of inter-individual variability would allow to improve the power of ICG based DLFTs and move toward an individualized evaluation of liver function. Within this work we systematically analyzed the effect of various factors on ICG elimination by the means of computational modeling. For the analysis, a recently developed and validated physiologically based pharmacokinetics (PBPK) model of ICG distribution and hepatic elimination was utilized. Key results are (i) a systematic analysis of the variability in ICG elimination due to hepatic blood flow, cardiac output, OATP1B3 abundance, liver volume, body weight and plasma bilirubin level; (ii) the evaluation of the inter-individual variability in ICG elimination via a large in silico cohort of n = 100,000 subjects based on the NHANES cohort with special focus on stratification by age, sex, and body weight; (iii) the evaluation of the effect of various degrees of cirrhosis on variability in ICG elimination. The presented results are an important step toward individualizing liver function tests by elucidating the effects of confounding physiological and anthropometric parameters in the evaluation of liver function via ICG.
Keywords:
ICG,
PBPK,
computational model,
indocyanine green,
liver cirrhosis,
liver function,
mathematical model,
pharmacokinetics,
|
|
| 2021 |


|
Hepatectomy-Induced Alterations in Hepatic Perfusion and Function - Toward Multi-Scale Computational Modeling for a Better Prediction of Post-hepatectomy Liver Function
Bruno Christ∗, Maximilian Collatz*, Uta Dahmen*, Karl-Heinz Herrmann*, Sebastian Höpfl*, Matthias König*, Lena Lambers*, Manja Marz*, Daria Meyer*, Nicole Radde*, Jürgen R. Reichenbach*, Tim Ricken*, and Hans-Michael Tautenhahn* (* equal contribution)
Front Physiol. 2021 Nov 18;12:733868. doi:10.3389/fphys.2021.733868. pmid:34867441
Liver resection causes marked perfusion alterations in the liver remnant both on the organ scale (vascular anatomy) and on the microscale (sinusoidal blood flow on tissue level). These changes in perfusion affect hepatic functions via direct alterations in blood supply and drainage, followed by indirect changes of biomechanical tissue properties and cellular function. Changes in blood flow impose compression, tension and shear forces on the liver tissue. These forces are perceived by mechanosensors on parenchymal and non-parenchymal cells of the liver and regulate cell-cell and cell-matrix interactions as well as cellular signaling and metabolism. These interactions are key players in tissue growth and remodeling, a prerequisite to restore tissue function after PHx. Their dysregulation is associated with metabolic impairment of the liver eventually leading to liver failure, a serious post-hepatectomy complication with high morbidity and mortality. Though certain links are known, the overall functional change after liver surgery is not understood due to complex feedback loops, non-linearities, spatial heterogeneities and different time-scales of events. Computational modeling is a unique approach to gain a better understanding of complex biomedical systems. This approach allows (i) integration of heterogeneous data and knowledge on multiple scales into a consistent view of how perfusion is related to hepatic function; (ii) testing and generating hypotheses based on predictive models, which must be validated experimentally and clinically. In the long term, computational modeling will (iii) support surgical planning by predicting surgery-induced perfusion perturbations and their functional (metabolic) consequences; and thereby (iv) allow minimizing surgical risks for the individual patient. Here, we review the alterations of hepatic perfusion, biomechanical properties and function associated with hepatectomy. Specifically, we provide an overview over the clinical problem, preoperative diagnostics, functional imaging approaches, experimental approaches in animal models, mechanoperception in the liver and impact on cellular metabolism, omics approaches with a focus on transcriptomics, data integration and uncertainty analysis, and computational modeling on multiple scales. Finally, we provide a perspective on how multi-scale computational models, which couple perfusion changes to hepatic function, could become part of clinical workflows to predict and optimize patient outcome after complex liver surgery.
Keywords:
liver perfusion,
liver regeneration,
liver surgery,
mechanoperception,
multi-scale modeling,
perfusion/function relationship,
rodent models of liver surgery,
|
|
| 2021 |


|
Specifications of Standards in Systems and Synthetic Biology: Status and Developments in 2021
F. Schreiber, P. Gleeson, M. Golebiewski, T. Gorochowski, M. Hucka, S. Keating, M. König, C. Myers, D. Nickerson, D. Walthemath
J Integr Bioinform. 2021 Oct 22;18(3):20210026. doi:10.1515/jib-2021-0026. pmid:34674411
This special issue of the Journal of Integrative Bioinformatics contains updated specifications of COMBINE standards in systems and synthetic biology. The 2021 special issue presents four updates of standards: Synthetic Biology Open Language Visual Version 2.3, Synthetic Biology Open Language Visual Version 3.0, Simulation Experiment Description Markup Language Level 1 Version 4, and OMEX Metadata specification Version 1.2. This document can also be consulted to identify the latest specifications of all COMBINE standards.
Keywords:
COMBINE,
standardization,
SBML,
SEDML,
OMEX,
|
|
| 2021 |


|
Computational modelling of omeprazole - pharmacokinetics and pharmacodynamics
Sükrü Balci (supervisor: Matthias König)
Bachelor Thesis, October 2021
Proton pump inhibitors (PPIs) are one of the most commonly used drugs whose main action is to inhibit the formation of gastric acid. Among the PPIs, omeprazole is one of the most important and most prescribed. Omeprazole is rapidly absorbed by the stomach and has a short half-life of around 1-2 hours in plasma. Omeprazole reduces gastric acid secretion via non-competitive selective inhibition of the H+/K+-ATPases in the gastric parietal cells, resulting in an increase in stomach pH. After cessation of omeprazole treatment, it takes a few days until the gastric acid secretion returns to baseline due to protein turnover of the proton pumps. Repeated dosing with omeprazole results in a dose-potentiation due to an increased absorption of omeprazole with increasing pH. Within this thesis, the pharmacokinetics and pharmacodynamics of omeprazole were studied using a computational modeling approach. Based on extensive curation of clinical data of omeprazole a physiological-based pharmacokinetics/pharmacodynamics model of omeprazole and its effect on stomach pH was developed. The model allowed to simulate the pharmacokinetics of omeprazole under varying doses and treatment regimens and to study the resulting inhibition of the proton pumps and changes in stomach pH. Key questions studied in this thesis were (i) how gastric acid output and pH change with omeprazole dosing and treatment regime, and ii) what the effects of repeated dosing on omeprazole absorption and pH are.
Keywords:
omeprazol,
PBPK,
pharmacokinetics,
pharmacodynamics,
proton pump inhibitor,
PPI,
|
|
| 2021 |



|
SBMLWebApp: Web-based Simulation, Steady-State Analysis, and Parameter Estimation of Systems Biology Models
Takahiro G. Yamada, Kaito Ii, Matthias König, Martina Feierabend, Andreas Dräger, and Akira Funahashi
Processes 9, no. 10: 1830. doi:10.3390/pr9101830
In systems biology, biological phenomena are often modeled by Ordinary Differential Equations (ODEs) and distributed in the de facto standard file format SBML. The primary analyses performed with such models are dynamic simulation, steady-state analysis, and parameter estimation. These methodologies are mathematically formalized, and libraries for such analyses have been published. Several tools exist to create, simulate, or visualize models encoded in SBML. However, setting up and establishing analysis environments is a crucial hurdle for non-modelers. Therefore, easy access to perform fundamental analyses of ODE models is a significant challenge. We developed SBMLWebApp, a web-based service to execute SBML-based simulation, steady-state analysis, and parameter estimation directly in the browser without the need for any setup or prior knowledge to address this issue. SBMLWebApp visualizes the result and numerical table of each analysis and provides a download of the results. SBMLWebApp allows users to select and analyze SBML models directly from the BioModels Database. Taken together, SBMLWebApp provides barrier-free access to an SBML analysis environment for simulation, steady-state analysis, and parameter estimation for SBML models. SBMLWebApp is implemented in Java™ based on an Apache Tomcat® web server using COPASI, the Systems Biology Simulation Core Library (SBSCL), and LibSBMLSim as simulation engines. SBMLWebApp is licensed under MIT with source code freely available. At the end of this article, the Data Availability Statement gives the internet links to the two websites to find the source code and run the program online.
Keywords:
SBML,
kinetic models,
time-course simulation,
steady-state simulation,
parameter estimation,
model calibration,
software,
web application,
|
|
| 2021 |



|
The simulation experiment description markup language (SED-ML): language specification for level 1 version 4
Smith, Lucian P., Bergmann, Frank T., Garny, Alan, Helikar, Tomáš, Karr, Jonathan, Nickerson, David, Sauro, Herbert, Waltemath, Dagmar and König, Matthias.
Journal of Integrative Bioinformatics, vol. 18, no. 3, 2021, pp. 20210021. doi:10.1515/jib-2021-0021. pmid:35330701
Computational simulation experiments increasingly inform modern biological research, and bring with them the need to provide ways to annotate, archive, share and reproduce the experiments performed. These simulations increasingly require extensive collaboration among modelers, experimentalists, and engineers. The Minimum Information About a Simulation Experiment (MIASE) guidelines outline the information needed to share simulation experiments. SED-ML is a computer-readable format for the information outlined by MIASE, created as a community project and supported by many investigators and software tools. The first versions of SED-ML focused on deterministic and stochastic simulations of models. Level 1 Version 4 of SED-ML substantially expands these capabilities to cover additional types of models, model languages, parameter estimations, simulations and analyses of models, and analyses and visualizations of simulation results. To facilitate consistent practices across the community, Level 1 Version 4 also more clearly describes the use of SED-ML constructs, and includes numerous concrete validation rules. SED-ML is supported by a growing ecosystem of investigators, model languages, and software tools, including eight languages for constraint-based, kinetic, qualitative, rule-based, and spatial models, over 20 simulation tools, visual editors, model repositories, and validators. Additional information about SED-ML is available at https://sed-ml.org/.
Keywords:
COMBINE,
SED-ML,
|
|
| 2021 |



|
The Systems Biology Simulation Core Library
Panchiwala, H.; Shah, S.; Planatscher, H.; Zakharchuk, M.; König, M.; Dräger, A.
Bioinformatics. 2022 Jan 12;38(3):864-865. doi:10.1093/bioinformatics/btab669. pmid:34554191
Summary: Studying biological systems generally relies on computational modeling and simulation, e.g., model-driven discovery and hypothesis testing. Progress in standardization efforts led to the development of interrelated file formats to exchange and reuse models in systems biology, such as SBML, the Simulation Experiment Description Markup Language (SED-ML) or the Open Modeling EXchange format. Conducting simulation experiments based on these formats requires efficient and reusable implementations to make them accessible to the broader scientific community and to ensure the reproducibility of the results. The Systems Biology Simulation Core Library (SBSCL) provides interpreters and solvers for these standards as a versatile open-source API in JavaTM. The library simulates even complex bio-models and supports deterministic Ordinary Differential Equations; Stochastic Differential Equations; constraint-based analyses; recent SBML and SED-ML versions; exchange of results, and visualization of in silico experiments; open modeling exchange formats (COMBINE archives); hierarchically structured models; and compatibility with standard testing systems, including the Systems Biology Test Suite and published models from the BioModels and BiGG databases.
|
|
| 2021 |

|
Ten Simple Rules for FAIR Sharing of Experimental and Clinical Data with the Modeling Community
Matthias König*, Jan Grzegorzewski, Martin Golebiewski, Henning Hermjakob, Mike Hucka, Brett Olivier, Sarah M. Keating, David Nickerson, Falk Schreiber, Rahuman Sheriff, Dagmar Waltemath
Preprints 2021, 2021080303 (in revision). doi:10.20944/preprints202108.0303.v2
Science continues to become more interdisciplinary and to involve increasingly complex data sets. Many projects in the biomedical and health-related sciences follow or aim to follow the principles of FAIR data sharing, which has been demonstrated to foster collaboration, to lead to better research outcomes, and to help ensure reproducibility of results. Data generated in the course of biomedical and health research present specific challenges for FAIR sharing in the sense that they are heterogeneous and highly sensitive to context and the needs of protection and privacy. Data sharing must respect these features without impeding timely dissemination of results, so that they can contribute to time-critical advances in medical therapy and treatment. Modeling and simulation of biomedical processes have become established tools, and a global community has been developing algorithms, methodologies, and standards for applying biomedical simulation models in clinical research. However, it can be difficult for clinician scientists to follow the specific rules and recommendations for FAIR data sharing within this domain. We seek to clarify the standard workflow for sharing experimental and clinical data with the simulation modeling community. By following these recommendations, data sharing will be improved, collaborations will become more effective, and the FAIR publication and subsequent reuse of data will become possible at the level of quality necessary to support biomedical and health-related sciences.
Keywords:
data sharing,
FAIR,
|
|
| 2021 |

|
OMEX Metadata Specification Version 1.2
John Gennari, Matthias König, Goskel Misirli, Maxwell Neal, David Nickerson, Dagmar Waltemath
J Integr Bioinform. 2021 Oct 20;18(3):20210020. doi:10.1515/jib-2021-0020. pmid:34668356
A standardized approach to annotating computational biomedical models and their associated files can facilitate model reuse and reproducibility among research groups, enhance search and retrieval of models and data, and enable semantic comparisons between models. Motivated by these potential benefits and guided by consensus across the COmputational Modeling in BIology NEtwork (COMBINE) community, we have developed a specification for encoding annotations in Open Modeling and EXchange (OMEX)-formatted archives. This document details version 1.2 of the specification, which builds on version 1.0 published last year in this journal. In particular, this version includes a set of initial model-level annotations (whereas v 1.0 described exclusively annotations at a smaller scale). Additionally, this version uses best practices for namespaces, and introduces omex-library.org as a common root for all annotations. Distributing modeling projects within an OMEX archive is a best practice established by COMBINE, and the OMEX metadata specification presented here provides a harmonized, community-driven approach for annotating a variety of standardized model representations. This specification acts as a technical guideline for developing software tools that can support this standard, and thereby encourages broad advances in model reuse, discovery, and semantic analyses.
|
|
| 2021 |



|
SED-ML Validator: tool for debugging simulation experiments
Bilal Shaikh, Andrew Philip Freiburger, Matthias König, Frank T. Bergmann, David P. Nickerson, Herbert M. Sauro, Michael L. Blinov, Lucian P. Smith, Ion I. Moraru, Jonathan R. Karr
arXiV, 2021, 2106.00844 (preprint, submitted). doi:10.48550/arXiv.2106.00844
|
|
| 2021 |

|
A Physiologically Based Model of Indocyanine Green Liver Function Tests - Effects of Physiological Factors, Hepatic Disease and Hepatic Surgery
Adrian Köller (supervisor: Matthias König)
Bachelor Thesis, March 2021
The evaluation of hepatic function and functional capacity are essential tasks in hepatology. Indocyanine Green (ICG) is a widely applied test compound that is used in clinical routine to evaluate hepatic function. Accurate assessment of liver-function through ICG pharmacokinetics is challenging because the elimination of ICG is influenced by physiological factors such as liver perfusion as well as the health status of the patient. Important questions for the functional evaluation are how blood flow, plasma protein binding or surgical interventions can influence the distribution and elimination of ICG. In the course of this project, these questions were studied by the means of computational modeling to evaluate ICG dynamics in silico. Within this work a physiological-based model of ICG pharmacokinetics was developed. For the parameterization and validation of the model a database of ICG pharmacokinetic data was established. The model was applied to study (i) the effect of liver disease, specifically liver cirrhosis, on ICG pharmacokinetics; (ii) how hepatic blood flow and other cardiovascular parameters affect ICG-elimination; and (iii) the role of plasma protein binding of ICG and its impact on ICG pharmacokinetics. Finally, the model was applied to analyse the effect of liver resection (hepatectomy) on ICG-elimination and the value of ICG as a predictive measure for postoperative outcome. The model is able to accurately predict changes in ICG pharmacokinetics caused by changes in blood flow and plasma proteins as well as liver cirrhosis. Furthermore, the model is able to accurately predict postoperative changes of ICG-elimination after liver resection, showing its potential value as a clinical tool.
|
|
| 2020 |


|
QuaLiPerF – Multi-X Liver Modelling
Tim Ricken, Lena Lambers, Bruno Christ, Uta Dahmen, Karl-Heinz Herrmann, Matthias König, Manja Marz, Nicole Radde, Jürgen R. Reichenbach, Lars Ole Schwen & Hans-Michael Tautenhahn
GACM - German Association for Computational Mechanics Report, winter 2020: No. 13
|
|
| 2020 |

|
Computational Modelling of Simvastatin - Effects on HMG-CoA Reductase Activity and Cholesterol
Florian Bartsch (supervisor: Matthias König)
Bachelor Thesis, November 2020
Cholesterol is one of the most important molecules in biology. Elevated plasma levels are associated with an increased risk for arteriosclerosis and cardiovascular diseases. A common treatment for hypercholesterolemic patients are statins, which are capable of reducing total plasma cholesterol levels. Within this thesis the effects of the statin simvastatin on cholesterol levels were studied using a computational modelling approach. Based on extensive data curation, a kinetic model of simvastatin pharmacokinetics coupled to a pharmacodynamic model of cholesterol metabolism was developed. The resulting pharmacokinetic/ pharmacodynamic (PK/PD) model was applied to study the effects of simvastatin on hepatic HMG-CoA reductase activity, hepatic cholesterol homeostasis and plasma cholesterol levels.
Keywords:
simvastaton,
PBPK,
pharmacokinetics,
pharmacodynamics,
cholesterol,
|
|
| 2020 |

|
Computational Modelling of Midazolam Clearance: Effect of Inhibitors and Inducers
Yannick Duport (supervisor: Matthias König)
Bachelor Thesis, August 2020
Midazolam is one of the most effective and safe medicines used for anesthesia and procedural sedation, as well as treating trouble sleeping and severe agitation. Midazolam is metabolised in the liver as well as in the small intestine to cytochrome P450 3A4 (CYP3A4). Many factors can alter the pharmacokinetics of midazolam due to inhibition or induction of hepatic and/or intestinal metabolism, resulting in either (i) adverse effects due to increased plasma midazolam concentrations in case of inhibition of metabolism or (ii) ineffectiveness as a consequence of too low concentrations if metabolism is induced. Within this thesis the clearance and pharmacokinetics of midazolam were studied, using a computational modeling approach. A large data base of midazolam pharmacokinetics data was established and utilized to develop a physiological-based pharmacokinetics model of midazolam. The model was applied to study the following questions: (i) What are the effects of CYP3A4 inhibition and induction on midazolam pharmacokinetics? (ii) What are the differences of intestinal and hepatic CYP3A4 inhibition and induction on midazolam pharmacokinetics?
Keywords:
midazolam,
PBPK,
pharmacokinetics,
drug-drug interactions,
DDI,
|
|
| 2020 |



|
PK-DB: pharmacokinetics database for individualized and stratified computational modeling
Grzegorzewski J, Brandhorst J, Green K, Eleftheriadou D, Duport Y, Barthorscht F, Köller A, Ke DYJ, De Angelis S, König M.
Nucleic Acids Res. 2021 Jan 8;49(D1):D1358-D1364. doi:10.1093/nar/gkaa990. pmid:33151297
A multitude of pharmacokinetics studies have been published. However, due to the lack of an open database, pharmacokinetics data, as well as the corresponding meta-information, have been difficult to access. We present PK-DB (https://pk-db.com), an open database for pharmacokinetics information from clinical trials. PK-DB provides curated information on (i) characteristics of studied patient cohorts and subjects (e.g. age, bodyweight, smoking status, genetic variants); (ii) applied interventions (e.g. dosing, substance, route of application); (iii) pharmacokinetic parameters (e.g. clearance, half-life, area under the curve) and (iv) measured pharmacokinetic time-courses. Key features are the representation of experimental errors, the normalization of measurement units, annotation of information to biological ontologies, calculation of pharmacokinetic parameters from concentration-time profiles, a workflow for collaborative data curation, strong validation rules on the data, computational access via a REST API as well as human access via a web interface. PK-DB enables meta-analysis based on data from multiple studies and data integration with computational models. A special focus lies on meta-data relevant for individualized and stratified computational modeling with methods like physiologically based pharmacokinetic (PBPK), pharmacokinetic/pharmacodynamic (PK/PD), or population pharmacokinetic (pop PK) modeling.
|
|
| 2020 |



|
SBML Level 3: an extensible format for the exchange and reuse of biological models
SM Keating, D Waltemath, M König, F Zhang, A Dräger, C Chaouiya, FT Bergmann, A Finney, CS Gillespie, T Helikar, S Hoops, RS Malik-Sheriff, SL Moodie, II Moraru, CJ Myers, A Naldi, BG Olivier, S Sahle, JC Schaff, LP Smith, MJ Swat, DT, L Watanabe, DJ Wilkinson, ML Blinov, K Begley, JR Faeder, HF Gómez, TM Hamm, Y Inagaki, W Liebermeister, AL Lister, D Lucio, E Mjolsness, CJ Proctor, K Raman, N Rodriguez, CA Shaffer, BE Shapiro, J Stelling, N Swainston, N Tanimura, J Wagner, M Meier-Schellersheim, HM Sauro, B Palsson, H Bolouri, H Kitano, Akira Funahashi, H Hermjakob, JC Doyle M Hucka, and SBML Community members
Mol Syst Biol. 2020;16(8):e9110. doi:10.15252/msb.20199110. pmid:32845085
Systems biology has experienced dramatic growth in the number, size, and complexity of computational models. To reproduce simulation results and reuse models, researchers must exchange unambiguous model descriptions. We review the latest edition of the Systems Biology Markup Language (SBML), a format designed for this purpose. A community of modelers and software authors developed SBML Level 3 over the past decade. Its modular form consists of a core suited to representing reaction-based models and packages that extend the core with features suited to other model types including constraint-based models, reaction-diffusion models, logical network models, and rule-based models. The format leverages two decades of SBML and a rich software ecosystem that transformed how systems biologists build and interact with models. More recently, the rise of multiscale models of whole cells and organs, and new data sources such as single-cell measurements and live imaging, has precipitated new ways of integrating data with models. We provide our perspectives on the challenges presented by these developments and how SBML Level 3 provides the foundation needed to support this evolution.
Keywords:
computational modeling,
file format,
interoperability,
reproducibility,
systems biology,
|
|
| 2020 |


|
Open modeling and exchange (OMEX) metadata specification version 1.0
Neal ML, Gennari JH, Waltemath D, Nickerson DP, König M
J Integr Bioinform. 2020 Jun 25;17(2-3):20200020. doi:10.1515/jib-2020-0020. pmid:32589606
A standardized approach to annotating computational biomedical models and their associated files can facilitate model reuse and reproducibility among research groups, enhance search and retrieval of models and data, and enable semantic comparisons between models. Motivated by these potential benefits and guided by consensus across the COmputational Modeling in BIology NEtwork (COMBINE) community, we have developed a specification for encoding annotations in Open Modeling and EXchange (OMEX)-formatted archives. Distributing modeling projects within these archives is a best practice established by COMBINE, and the OMEX metadata specification presented here provides a harmonized, community-driven approach for annotating a variety of standardized model and data representation formats within an archive. The specification primarily includes technical guidelines for encoding archive metadata, so that software tools can more easily utilize and exchange it, thereby spurring broad advancements in model reuse, discovery, and semantic analyses.
Keywords:
data integration,
metadata,
semantics,
simulation,
|
|
| 2020 |


|
Specifications of Standards in Systems and Synthetic Biology:Status and Developments in 2020
F. Schreiber, B. Sommer, G. Bader, P. Gleeson, M. Golebiewski, T. Gorochowski, M. Hucka, S. Keating, M. König, C. Myers, D. Nickerson, D. Walthemath
J Integr Bioinform. 2020 Jun 29;17(2-3):20200022. doi:10.1515/jib-2020-0022
This special issue of the Journal of Integrative Bioinformatics presents papers related to the 10th COMBINE meeting together with the annual update of COMBINE standards in systems and synthetic biology.
Keywords:
ontologies,
standards,
synthetic biology,
systems biology,
|
|
| 2020 |


|
The first 10 years of the international coordination network for standards in systems and synthetic biology (COMBINE)
D Waltemath, M Golebiewski, M Blinov, P Gleeson, H Hermjakob, E Inau, S Keating, M König, O Krebs, R Malik-Sheriff, D Nickerson, E Oberortner, H Sauro, F Schreiber, L Smith, M Stefan, U Wittig, C Myers
J Integr Bioinform. 2020 Jun 29;17(2-3):20200005. doi:10.1515/jib-2020-0005
This paper presents a report on outcomes of the 10th Computational Modeling in Biology Network (COMBINE) meeting that was held in Heidelberg, Germany, in July of 2019. The annual event brings together researchers, biocurators and software engineers to present recent results and discuss future work in the area of standards for systems and synthetic biology. The COMBINE initiative coordinates the development of various community standards and formats for computational models in the life sciences. Over the past 10 years, COMBINE has brought together standard communities that have further developed and harmonized their standards for better interoperability of models and data. COMBINE 2019 was co-located with a stakeholder workshop of the European EU-STANDS4PM initiative that aims at harmonized data and model standardization for in silico models in the field of personalized medicine, as well as with the FAIRDOM PALs meeting to discuss findable, accessible, interoperable and reusable (FAIR) data sharing. This report briefly describes the work discussed in invited and contributed talks as well as during breakout sessions. It also highlights recent advancements in data, model, and annotation standardization efforts. Finally, this report concludes with some challenges and opportunities that this community will face during the next 10 years.
Keywords:
COMBINE,
community building,
meeting report,
standardization,
|
|
| 2020 |


|
The Distributions Package for SBML Level 3
L Smith, S Moodie, F Bergmann, C Gillespie, S Keating, M. König, C Myers, M Swat, D Wilkinson, M Hucka
J Integr Bioinform. 2020 Jul 20;17(2-3):20200018. doi:10.1515/jib-2020-0018. pmid:32750035
Biological models often contain elements that have inexact numerical values, since they are based on values that are stochastic in nature or data that contains uncertainty. The Systems Biology Markup Language (SBML) Level 3 Core specification does not include an explicit mechanism to include inexact or stochastic values in a model, but it does provide a mechanism for SBML packages to extend the Core specification and add additional syntactic constructs. The SBML Distributions package for SBML Level 3 adds the necessary features to allow models to encode information about the distribution and uncertainty of values underlying a quantity.
Keywords:
SBML,
distributions,
modeling,
systems biology,
uncertainty,
|
|
| 2020 |



|
MEMOTE for standardized genome-scale metabolic model testing
Christian Lieven, Moritz Emanuel Beber, Brett G. Olivier, Frank T. Bergmann, Meric Ataman, Parizad Babaei, Jennifer A. Bartell, Lars M. Blank, Siddharth Chauhan, Kevin Correia, Christian Diener, Andreas Dräger, Birgitta Elisabeth Ebert, Janaka N. Edirisinghe, Jose P. Faria, Adam Feist, Georgios Fengos, Ronan M. T. Fleming, Beatriz Garcia-Jimenez, Vassily Hatzimanikatis, Wout van Helvoirt, Christopher Henry, Henning Hermjakob, Markus J. Herrgard, Hyun Uk Kim, Zachary King, Jasper Jan Koehorst, Matthias König, Steffen Klamt, Edda Klipp, Meiyappan Lakshmanan, Nicolas Le Novere, Dong-Yup Lee, Sang Yup Lee, Sunjae Lee, Nathan E. Lewis, Hongwu Ma, Daniel Machado, Radhakrishnan Mahadevan, Paulo Maia, Adil Mardinoglu, Greg L. Medlock, Jonathan Monk, Jens Nielsen, Lars K. Nielsen, Juan Nogales, Intawat Nookaew, Osbaldo Resendis, Bernhard Palsson, Jason A. Papin, Kiran Raosaheb Patil, Mark Poolman, Nathan D. Price, Anne Richelle, Isabel Rocha, Benjamin Sanchez, Peter Schaap, Rahuman S. Malik Sheriff, Saeed Shoaie, Nikolaus Sonnenschein, Bas Teusink, Paulo Vilaca, Jon Olav Vik, Judith A. Wodke, Joana C. Xavier, Qianqian Yuan, Maksim Zakhartsev, Cheng Zhang
Nat Biotechnol. 2020;38(3):272-276. doi:10.1038/s41587-020-0446-y. pmid:32123384
Reconstructing metabolic reaction networks enables the development of testable hypotheses of an organism’s metabolism under different conditions. State-of-the-art genome-scale metabolic models (GEMs) can include thousands of metabolites and reactions that are assigned to subcellular locations. Gene–protein–reaction (GPR) rules and annotations using database information can add meta-information to GEMs. GEMs with metadata can be built using standard reconstruction protocols, and guidelines have been put in place for tracking provenance and enabling interoperability, but a standardized means of quality control for GEMs is lacking. Here we report a community effort to develop a test suite named MEMOTE (for metabolic model tests) to assess GEM quality.
|
|
| 2020 |



|
Executable Simulation Model of the Liver
Matthias König
bioRxiv 2020.01.04.894873 (preprint). doi:10.1101/2020.01.04.894873v1
To address the issue of reproducibility in computational modeling we developed the concept of an executable simulation model (EXSIMO). An EXSIMO combines model, data and code with the execution environment to run the computational analysis in an automated manner using tools from software engineering. Key components are i) models, data and code for the computational analysis; ii) tests for models, data and code; and iii) an automation layer to run tests and execute the analysis. An EXSIMO combines version control, model, data, units, annotations, analysis, reports, execution environment, testing, continuous integration and release. We applied the concept to perform a replication study of a computational analysis of hepatic glucose metabolism in the liver. The corresponding EXSIMO is available from https://github.com/matthiaskoenig/exsimo
|
|
| 2019 |

|
Model Order Reduction (MOR) of Function-Perfusion-Growth Simulation in the Human Fatty Liver via Artificial Neural Network (ANN)
Lambers L., Ricken T., and König M.
Proceedings in Applied Mathematics & Mechanics, PAMM, 2019. doi:10.1002/pamm.201900429
Numerical modeling of biological systems has become an important assistance for understanding and predicting hepatic diseases like non-alcoholic fatty liver disease (NAFLD) or the detoxification of drugs and toxines by the liver. We developed a model for the simulation of hepatic function-perfusion processes using a multiscale and multiphase approach. Here, the liver lobules are described using a homogenization approach with a coupled set of partial differential equations (PDE) based on the Theory of Porous Media (TPM) to describe the coupled blood transport and tissue deformation. For the description of metabolic processes on cellular scale ordinary differential equations (ODE) are used. For many practical and clinical applications, e.g. optimization procedures or uncertainty quantification, a fast but reliable computation is required. Thus, we use a non-linear model order reduction (MOR) based on an artificial neural network (ANN) for the prediction of simulation results. The practicability of this approach is shown in a comparison between the high fidelity numerical simulation of a NAFLD and the predicted results by the ANN.
|
|
| 2019 |


|
Specifications of Standards in Systems and Synthetic Biology: Status and Developments in 2019
F Schreiber, G Bader, P Gleeson, M Golebiewski, M Hucka, SM Keating, M König, C Myers, D Nickerson, B Sommer, D Waltemath
J Integr Bioinform. 2019 Jul 13;16(2). doi:10.1515/jib-2019-0035. pmid:31301675
This special issue of the Journal of Integrative Bioinformatics presents an overview of COMBINE standards and their latest specifications. The standards cover representation formats for computational modeling in synthetic and systems biology and include BioPAX, CellML, NeuroML, SBML, SBGN, SBOL and SED-ML. The articles in this issue contain updated specifications of SBGN Process Description Level 1 Version 2, SBML Level 3 Core Version 2 Release 2, SBOL Version 2.3.0, and SBOL Visual Version 2.1.
|
|
| 2019 |



|
The Systems Biology Markup Language (SBML): Language Specification for Level 3 Version 2 Core
M. Hucka, F. Bergmann, C. Chaouiya, A. Dräger, S. Hoops, S. Keating, M. König, N Le Novere, C. Myers, B. Olivier, S. Sahle, J. Schaff, R. Sheriff, L. Smith, D. Waltemath, D. Wilkinson, F. Zhang
J Integr Bioinform. 2019 Jun 20;16(2);. doi:10.1515/jib-2019-0021. pmid:31219795
Computational models can help researchers to interpret data, understand biological functions, and make quantitative predictions. The Systems Biology Markup Language (SBML) is a file format for representing computational models in a declarative form that different software systems can exchange. SBML is oriented towards describing biological processes of the sort common in research on a number of topics, including metabolic pathways, cell signaling pathways, and many others. By supporting SBML as an input/output format, different tools can all operate on an identical representation of a model, removing opportunities for translation errors and assuring a common starting point for analyses and simulations. This document provides the specification for Release 2 of Version 2 of SBML Level 3 Core. The specification defines the data structures prescribed by SBML as well as their encoding in XML, the eXtensible Markup Language. Release 2 corrects some errors and clarifies some ambiguities discovered in Release 1. This specification also defines validation rules that determine the validity of an SBML document, and provides many examples of models in SBML form. Other materials and software are available from the SBML project website at http://sbml.org/.
Keywords:
Representation,
Standards,
Systems Biology Markup Language,
Visualization,
|
|
| 2018 |



|
Tellurium: An Extensible Python-based Modeling Environment for Systems and Synthetic Biology
Choi K., Medley K., König M., Stocking K., Smith L., Gu S., and Sauro HM.
Biosystems. 2018 Jul 24. pii: S0303-2647(18)30125-4. doi:10.1016/j.biosystems.2018.07.006. pmid:30053414
Here we present Tellurium, a Python-based environment for model building, simulation, and analysis that facilitates reproducibility of models in systems and synthetic biology. Tellurium is a modular, cross-platform, and open-source simulation environment composed of multiple libraries, plugins, and specialized modules and methods. Tellurium is a self-contained modeling platform which comes with a fully configured Python distribution. Two interfaces are provided, one based on the Spyder IDE which has an accessible user interface akin to MATLAB and a second based on the Jupyter Notebook, which is a format that contains live code, equations, visualizations and narrative text. Tellurium uses libRoadRunner as the default SBML simulation engine which supports deterministic simulations, stochastic simulations, and steady-state analyses. Tellurium also includes Antimony, a human-readable model definition language which can be converted to and from SBML. Other standard Python scientific libraries such as NumPy, SciPy, and matplotlib are included by default. Additionally, we include several user-friendly plugins and advanced modules for a wide-variety of applications, ranging from complex algorithms for bifurcation analysis to multidimensional parameter scanning. By combining multiple libraries, plugins, and modules into a single package, Tellurium provides a unified but extensible solution for biological modeling and analysis for both novices and experts.
|
|
| 2018 |

|
HEPATOKIN1: A Biochemistry-Based Model of Liver Metabolism Suited for Applications in Medicine and Pharmacology
Berndt N., Bulik S., Wallach I., Wünsch T., König M., Stockmann M., Meierhofer M., Holzhütter HG.
Nat Commun. 2018 Jun 19;9(1):2386.. doi:10.1038/s41467-018-04720-9. pmid:29921957
The most reliable and predictive type of metabolic models are biochemistry-based kinetic models resting on the principles of chemical kinetics and including all relevant molecular details of enzyme regulation. Here we present such a model with yet unequalled complexity encompassing the major pathways of hepatic energy, carbohydrate, lipid, and nitrogen metabolism and including the regulation of enzyme activities by their reactants, allosteric effectors and hormone-dependent reversible enzyme phosphorylation. We illustrate the utility of the model for basic research and applications in medicine and pharmacology by simulating diurnal variations of the metabolic state of the liver under normal conditions and at various perturbations caused by nutritional challenges (alcohol), drugs (valproate) and inherited enzyme disorders (galactosemia). In particular, the model can be used to quantify alterations in the metabolic functions of different liver cells (normal hepatocytes versus adenoma and hepatocellular carcinoma) by exploiting relative changes in the abundance of metabolic enzymes for the re-scaling of maximal enzyme activities (Vmax values).
|
|
| 2018 |



|
Simulation Experiment Description Markup Language (SED-ML) Level 1 Version 3 (L1V3)
Bergmann, FT., Cooper, J., König, M.*, Moraru, I., Nickerson, D., Le Novère N., Olivier, BG., Sahle, S., Smith, L., Waltemath, D.
* corresponding author
J Integr Bioinform. 2018 Mar 19;15(1). doi:10.1515/jib-2017-0086. pmid:29550789
The creation of computational simulation experiments to inform modern biological research poses challenges to reproduce, annotate, archive, and share such experiments. Efforts such as SBML or CellML standardize the formal representation of computational models in various areas of biology. The Simulation Experiment Description Markup Language (SED-ML) describes what procedures the models are subjected to, and the details of those procedures. These standards, together with further COMBINE standards, describe models sufficiently well for the reproduction of simulation studies among users and software tools. The Simulation Experiment Description Markup Language (SED-ML) is an XML-based format that encodes, for a given simulation experiment, i) which models to use; ii) which modifications to apply to models before simulation; iii) which simulation procedures to run on each model; iv) how to post-process the data; and v) how these results should be plotted and reported. SED-ML Level 1 Version 1 (L1V1) implemented support for the encoding of basic time course simulations. SED-ML L1V2 added support for more complex types of simulations, specifically repeated tasks and chained simulation procedures. SED-ML L1V3 extends L1V2 by means to describe which datasets and subsets thereof to use within a simulation experiment.
|
|
| 2018 |



|
Harmonizing semantic annotations for computational models in biology
Neal, M.; König, M.; Nickerson, D.; Misirli, G.; Kalbasi, R.; Dräger, A.; Atalag, K.; Chelliah, V.; Cooling, M.; Cook, D.; Crook, S.; de Alba, M.; Friedman, S.; Garny, A.; Gennari, J.; Gleeson, P.; Golebiewski, M.; Hucka, M.; Juty, N.; Novère, N. L.; Myers, C.; Olivier, B.; Sauro, H.; Scharm, M.; Snoep, J.; Touré, V.; Wipat, A.; Wolkenhauer, O. & Waltemath, D.
Brief Bioinform. 2019 Mar 22;20(2):540-550. doi:10.1093/bib/bby087. pmid:30462164
Life science researchers use computational models to articulate and test hypotheses about the behavior of biological systems. Semantic annotation is a critical component for enhancing the interoperability and reusability of such models as well as for the integration of the data needed for model parameterization and validation. Encoded as machine-readable links to knowledge resource terms, semantic annotations describe the computational or biological meaning of what models and data represent. These annotations help researchers find and repurpose models, accelerate model composition, and enable knowledge integration across model repositories and experimental data stores. However, realizing the potential benefits of semantic annotation requires the development of model annotation standards that adhere to a community-based annotation protocol. Without such standards, tool developers must account for a variety of annotation formats and approaches, a situation that can become prohibitively cumbersome and which can defeat the purpose of linking model elements to controlled knowledge resource terms. Currently, no consensus protocol for semantic annotation exists among the larger biological modeling community. Here, we report on the landscape of current semantic annotation practices among the COmputational Modeling in BIology NEtwork (COMBINE) community and provide a set of recommendations for building a consensus approach to semantic annotation.
|
|
| 2018 |



|
Tellurium Notebooks — An Environment for Reproducible Dynamical Modeling in Systems Biology.
Medley K., Choi K., König M., Smith L., Gu S., Hellerstein J., Sealfon S., and Sauro HM.
PLoS Comput Biol. 2018 Jun 15;14(6):e1006220. doi:10.1371/journal.pcbi.1006220. pmid:29906293
The considerable difficulty encountered in reproducing the results of published dynamical models limits validation, exploration and reuse of this increasingly large biomedical research resource. To address this problem, we have developed Tellurium Notebook, a software system that facilitates building reproducible dynamical models and reusing models by 1) supporting the COMBINE archive format during model development for capturing model information in an exchangeable format and 2) enabling users to easily simulate and edit public COMBINE-compliant models from public repositories to facilitate studying model dynamics, variants and test cases. Tellurium Notebook, a Python-based Jupyter-like environment, is designed to seamlessly inter-operate with these community standards by automating conversion between COMBINE standards formulations and corresponding in-line, human-readable representations. Thus, Tellurium brings to systems biology the strategy used by other literate notebook systems such as Mathematica. These capabilities allow users to edit every aspect of the standards-compliant models and simulations, run the simulations in-line, and re-export to standard formats. We provide several use cases illustrating the advantages of our approach and how it allows development and reuse of models without requiring technical knowledge of standards. Adoption of Tellurium should accelerate model development, reproducibility and reuse.
|
|
| 2017 |

|
Computational Modeling in Liver Surgery
Christ, B.*, Dahmen, U.*, Herrmann, K.H.*, König, M.*, Reichenbach, J.R*., Ricken, T.*, Schleicher, J.*, Ole Schwen, L.*, Vlaic, S.* and Waschinsky, N.*
* equal contribution
Front Physiol. 2017 Nov 14;8:906. doi:10.3389/fphys.2017.00906. pmid:29249974
The need for extended liver resection is increasing due to the growing incidence of liver tumors in aging societies. Individualized surgical planning is the key for identifying the optimal resection strategy and to minimize the risk of postoperative liver failure and tumor recurrence. Current computational tools provide virtual planning of liver resection by taking into account the spatial relationship between the tumor and the hepatic vascular trees, as well as the size of the future liver remnant. However, size and function of the liver are not necessarily equivalent. Hence, determining the future liver volume might misestimate the future liver function, especially in cases of hepatic comorbidities such as hepatic steatosis. A systems medicine approach could be applied, including biological, medical, and surgical aspects, by integrating all available anatomical and functional information of the individual patient. Such an approach holds promise for better prediction of postoperative liver function and hence improved risk assessment. This review provides an overview of mathematical models related to the liver and its function and explores their potential relevance for computational liver surgery. We first summarize key facts of hepatic anatomy, physiology, and pathology relevant for hepatic surgery, followed by a description of the computational tools currently used in liver surgical planning. Then we present selected state-of-the-art computational liver models potentially useful to support liver surgery. Finally, we discuss the main challenges that will need to be addressed when developing advanced computational planning tools in the context of liver surgery.
Keywords:
Liver resection,
function prediction,
liver metabolism,
liver regeneration,
liver surgical planning,
multi-scale modeling,
risk assessment,
systems medicine,
|
|
| 2016 |

|
Challenges and opportunities for system biology standards and tools in medical research
König M.*, Oellrich A.*, Waltemath D.*, Dobson R., Hubbard T., and Wolkenhauer O.
* equal contribution
ODLS, p1-6, 2016; article
Kinetic models are increasingly relevant in medical research. In systems biology, more than 10 years of experience with the development of standards and tools to construct and analyse kinetic models exists. This has supported the sharing of kinetic models, increased their reuse, and thereby has helped to reproduce and validate scientific results. Given the expertise and the existing infrastructure, it seems natural to consider the application and development of standards and tools to meet the requirements of medical scientists. In this paper, we discuss challenges and opportunities for standards and tools from systems biology in medical research, and we put forward criteria for the safe use of simulations. We conclude that standards, tools and infrastructure need to be extended to ensure the quality, reliability and safety required when working with medical and patient data. This will foster the adaptation of modelling in the clinic, providing tools for improved diagnosis, prognosis and therapy.
|
|
| 2016 |



|
cy3sabiork: A Cytoscape app for visualizing kinetic data from SABIO-RK
König M.
[version 1; referees: awaiting peer review]. F1000Research 2016, 5:1736. doi:10.12688/f1000research.9211.1
Kinetic data of biochemical reactions are essential for the creation of kinetic models of biochemical networks. One of the main resources of such information is SABIO-RK, a curated database for kinetic data of biochemical reactions and their related information. Despite the importance for computational modelling there has been no simple solution to visualize the kinetic data from SABIO-RK. In this work, I present cy3sabiork, an app for querying and visualization of kinetic data from SABIO-RK in Cytoscape. The kinetic information is accessible via a combination of graph structure and annotations of nodes, with provided information consisting of: (I) reaction details, enzyme and organism; (II) kinetic law, formula, parameters; (III) experimental conditions; (IV) publication; (V) additional annotations. cy3sabiork creates an intuitive visualization of kinetic entries in form of a species-reaction-kinetics graph, which reflects the reaction-centered approach of SABIO-RK. Kinetic entries can be imported in SBML format from either the SABIO-RK web interface or via web service queries. The app allows for easy comparison of kinetic data, visual inspection of the elements involved in the kinetic record and simple access to the annotation information of the kinetic record. I applied cy3sabiork in the computational modelling of galactose metabolism in the human liver.
|
|
| 2016 |



|
Toward community standards and software for whole-cell modeling
Wholecell Consortium (König M.)
IEEE Trans Biomed Eng. 2016 Jun 10.. doi:10.1109/TBME.2016.2560762. pmid:27305665
Objective: Whole-cell (WC) modeling is a promising tool for biological research, bioengineering, and medicine. However, substantial work remains to create accurate comprehensive models of complex cells. Methods: We organized the 2015 Whole-Cell Modeling Summer School to teach WC modeling and evaluate the need for new WC modeling standards and software by recoding a recently published WC model in the Systems Biology Markup Language. Results: Our analysis revealed several challenges to representing WC models using the current standards. Conclusion: We, therefore, propose several new WC modeling standards, software, and databases. Significance: We anticipate that these new standards and software will enable more comprehensive models.
|
|
| 2015 |



|
Pathobiochemical signatures of cholestatic liver disease in bile duct ligated mice
Kerstin A.*, König M.*, Hoppe A., Thomas M., Müller I., Ebert M., Weng H., Holzhütter HG., Zanger UM., Bode J., Vollmar B. and Dooley S.
* equal contribution
BMC Syst Biol. 2015 Nov 20;9(1):83.. doi:10.1186/s12918-015-0229-0. pmid:26589287
Background: Disrupted bile secretion leads to liver damage characterized by inflammation, fibrosis, eventually cirrhosis, and hepatocellular cancer. As obstructive cholestasis often progresses insidiously, markers for the diagnosis and staging of the disease are urgently needed. To this end, we compiled a comprehensive data set of serum markers, histological parameters and transcript profiles at 8 time points of disease progression after bile duct ligation (BDL) in mice, aiming at identifying a set of parameters that could be used as robust biomarkers for transition of different disease progression phases. Results: Statistical analysis of the more than 6,000 data points revealed distinct temporal phases of disease. Time course correlation analysis of biochemical, histochemical and mRNA transcript parameters (=factors) defined 6 clusters for different phases of disease progression. The number of CTGF-positive cells provided the most reliable overall measure for disease progression at histological level, bilirubin at biochemical level, and metalloproteinase inhibitor 1 (Timp1) at transcript level. Prominent molecular events exhibited by strong transcript peaks are found for the transcriptional regulator Nr0b2 (Shp) and 1,25-dihydroxyvitamin D(3) 24-hydroxylase (Cyp24a1) at 6 h. Based on these clusters, we constructed a decision tree of factor combinations potentially useful as markers for different time intervals of disease progression. Best prediction for onset of disease is achieved by fibronectin (Fn1), for early disease phase by Cytochrome P450 1A2 (Cyp1a2), passage to perpetuation phase by collagen1α-1 (Col1a1), and transition to the progression phase by interleukin 17-a (Il17a), with early and late progression separated by Col1a1. Notably, these predictions remained stable even for randomly chosen small sub-sets of factors selected from the clusters. Conclusion: Our detailed time-resolved explorative study of liver homogenates following BDL revealed a well-coordinated response, resulting in disease phase dependent parameter modulations at morphological, biochemical, metabolic and gene expression levels. Interestingly, a small set of selected parameters can be used as diagnostic markers to predict disease stages in mice with cholestatic liver disease.
|
|
| 2015 |
|
On the Influence of Growth in Perfusion Dependent Biological Systems - at the Example of the Human Liver.
Werner D., Ricken T., Dahmen U., Dirsch O., Holzhütter HG., König M.
PAMM 15 (1), 119-120, 2015. doi:10.1002/pamm.201510050
Biochemical reactions in the liver are a complex non-linear and time depending coupled function-perfusion-mechanisms. The metabolism of the liver, e.g. nutrient storage, is directly coupled to its blood perfusion. Growth phenomena, such as accumulation of fat, have a strong impact onto micro-perfusion within the liver. Here, we present the results of a computational model that describes selected coupled functionalities. The metabolic processes take place in the liver cells, the hepatocytes, which are arranged in a delicate system of capillaries, so called sinusoids. Nutrients, oxygen, and other components of the blood are transported through the sinusoids. Due to this highly complex inner structure of the lobules it is impracticable to give a microscopic geometrical description in a continuum mechanical manner. Therefore, a homogenization scheme based on the theory of porous media (TPM) is used;. In case of the presented liver model we consider a porous solid body, that consists of tissue and fat, and a fluid representing blood. In each phase several miscible components are considered. For the microscopic cell level use has been made of an embedded set of ordinary differential equations (ODE) that mimics a simplified metabolism, e.g. glucose utilization and production;. In case of growth of the fat phase we follow a phenomenological approach. Numerical examples for this coupled process are presented and discussed.
|
|
| 2015 |



|
LibRoadRunner: a high performance SBML simulation and analysis library.
Somogyi ET., Bouteiller JM., Glazier JA., König M., Medley JK., Swat MH and Sauro HM.
Bioinformatics. 2015 Oct 15;31(20):3315-21. doi:10.1093/bioinformatics/btv363. pmid:26085503
Motivation: This article presents libRoadRunner, an extensible, high-performance, cross-platform, open-source software library for the simulation and analysis of models expressed using Systems Biology Markup Language (SBML). SBML is the most widely used standard for representing dynamic networks, especially biochemical networks. libRoadRunner is fast enough to support large-scale problems such as tissue models, studies that require large numbers of repeated runs and interactive simulations. Results: libRoadRunner is a self-contained library, able to run both as a component inside other tools via its C++ and C bindings, and interactively through its Python interface. Its Python Application Programming Interface (API) is similar to the APIs of MATLAB and SciPy, making it fast and easy to learn. libRoadRunner uses a custom Just-In-Time (JIT) compiler built on the widely used LLVM JIT compiler framework. It compiles SBML-specified models directly into native machine code for a variety of processors, making it appropriate for solving extremely large models or repeated runs. libRoadRunner is flexible, supporting the bulk of the SBML specification (except for delay and non-linear algebraic equations) including several SBML extensions (composition and distributions). It offers multiple deterministic and stochastic integrators, as well as tools for steady-state analysis, stability analysis and structural analysis of the stoichiometric matrix. Availability and implementation: libRoadRunner binary distributions are available for Mac OS X, Linux and Windows. The library is licensed under Apache License Version 2.0. libRoadRunner is also available for ARM-based computers such as the Raspberry Pi. http://www.libroadrunner.org provides online documentation, full build instructions, binaries and a git source repository.
|
|
| 2014 |

|
Modeling function-perfusion behavior in liver lobules including tissue, blood, glucose, lactate and glycogen by use of a coupled two-scale PDE-ODE approach.
Ricken T., Werner D., Holzhütter HG., König M., Dahmen U., Dirsch O.
Biomech Model Mechanobiol. 2015 Jun;14(3):515-36. doi:10.1007/s10237-014-0619-z. pmid:25236798
This study focuses on a two-scale, continuum multicomponent model for the description of blood perfusion and cell metabolism in the liver. The model accounts for a spatial and time depending hydro-diffusion-advection-reaction description. We consider a solid-phase (tissue) containing glycogen and a fluid-phase (blood) containing glucose as well as lactate. The five-component model is enhanced by a two-scale approach including a macroscale (sinusoidal level) and a microscale (cell level). The perfusion on the macroscale within the lobules is described by a homogenized multiphasic approach based on the theory of porous media (mixture theory combined with the concept of volume fraction). On macro level, we recall the basic mixture model, the governing equations as well as the constitutive framework including the solid (tissue) stress, blood pressure and solutes chemical potential. In view of the transport phenomena, we discuss the blood flow including transverse isotropic permeability, as well as the transport of solute concentrations including diffusion and advection. The continuum multicomponent model on the macroscale finally leads to a coupled system of partial differential equations (PDE). In contrast, the hepatic metabolism on the microscale (cell level) was modeled via a coupled system of ordinary differential equations (ODE). Again, we recall the constitutive relations for cell metabolism level. A finite element implementation of this framework is used to provide an illustrative example, describing the spatial and time-depending perfusion-metabolism processes in liver lobules that integrates perfusion and metabolism of the liver.
|
|
| 2014 |

|
Homeostasis of blood glucose - Computer simulations of central liver functions.
König M. and Holzhütter HG.
systembiologie.de 2014; 8:p.53-57 [PDF (de), PDF (en)]
The liver performs a multitude of important physiological functions such as synthesising plasma proteins, producing bile and hormones, detoxifying toxic compounds and precisely regulating the concentration of various substances in the blood (homeostasis). In particular, the liver plays a crucial role in the regulation of blood glucose levels. Modelling hepatic glucose metabolism helps to understand central liver functions and basic regulation mechanisms and thereby provides insights in the role of the liver in systemic diseases such as diabetes.
|
|
| 2013 |

|
Metabolic Gradients as Key Regulators in Zonation of Tumor Energy Metabolism: A Tissue-scale Model Based Study.
König M., Holzhütter HG., Berndt N.
Biotechnol J. 2013 Sep;8(9):1058-69.. doi:10.1002/biot.201200393. pmid:23589477
Characteristics of many tumor types are the reprogramming of metabolism and the occurrence of regional hypoxia. In this work, we investigated the hypothesis that metabolic reprogramming in combination with metabolic zonation of cellular energy metabolism are important factors in promotion of the growth capacity of solid tumors. A tissue-scale model of the two main ATP delivering pathways, glycolysis (GLY) and oxidative phosphorylation (OXP), was used to simulate the energy metabolism within solid tumors under various metabolic strategies. Remarkably, despite the high diversity in the usage of glucose, lactate and oxygen in various spatial regions, the tumor as a whole clearly displays the hallmark of the so-called Warburg effect, i.e. a high rate of glucose consumption and lactate production in the presence of sufficiently high levels of oxygen. Our simulations suggest that an increase in GLY capacity and concomitant decrease in OXP capacity from the periphery towards the center of the tumor improves the availability of oxygen to pericentral tumor cells. The found relationship between the regional oxygen level and the relative share of GLY and OXP capacities supports the view that metabolite availability functions as key regulator of tumor energy metabolism.
|
|
| 2012 |

|
Kinetic Modeling of Human Hepatic Glucose Metabolism in T2DM Predicts Higher Risk of Hypoglycemic Events in Rigorous Insulin Therapy
König M. and Holzhütter HG.
J Biol Chem. 2012 Oct 26;287(44):36978-89. doi:10.1074/jbc.M112.382069. pmid:22977253
A major problem in the insulin therapy of patients with diabetes type 2 (T2DM) is the increased occurrence of hypoglycemic events which, if left untreated, may cause confusion or fainting and in severe cases seizures, coma, and even death. To elucidate the potential contribution of the liver to hypoglycemia in T2DM we applied a detailed kinetic model of human hepatic glucose metabolism to simulate changes in glycolysis, gluconeogenesis, and glycogen metabolism induced by deviations of the hormones insulin, glucagon, and epinephrine from their normal plasma profiles. Our simulations reveal in line with experimental and clinical data from a multitude of studies in T2DM, (i) significant changes in the relative contribution of glycolysis, gluconeogenesis, and glycogen metabolism to hepatic glucose production and hepatic glucose utilization; (ii) decreased postprandial glycogen storage as well as increased glycogen depletion in overnight fasting and short term fasting; and (iii) a shift of the set point defining the switch between hepatic glucose production and hepatic glucose utilization to elevated plasma glucose levels, respectively, in T2DM relative to normal, healthy subjects. Intriguingly, our model simulations predict a restricted gluconeogenic response of the liver under impaired hormonal signals observed in T2DM, resulting in an increased risk of hypoglycemia. The inability of hepatic glucose metabolism to effectively counterbalance a decline of the blood glucose level becomes even more pronounced in case of tightly controlled insulin treatment. Given this Janus face mode of action of insulin, our model simulations underline the great potential that normalization of the plasma glucagon profile may have for the treatment of T2DM.
|
|
| 2012 |



|
CySBML: a Cytoscape plugin for SBML
König M., Dräger A. and Holzhütter HG.
Bioinformatics. 2012 Sep 15;28(18):2402-3. doi:10.1093/bioinformatics/bts432. pmid:22772946
Summary: CySBML is a plugin designed to work with Systems Biology Markup Language (SBML) in Cytoscape having the following features: SBML import, support of the SBML layout and qualitative model packages, navigation in network layouts based on SBML structure, access to MIRIAM and SBO-based annotations and SBML validation. CySBML includes an importer for BioModels to load SBML from standard repositories. Availability and implementation: Freely available for non-commercial purposes through the Cytoscape plugin manager or for download at http://sourceforge.net/projects/cysbml/. Contact: cysbml-team@lists.sourceforge.net. Supplementary information: Tutorial, usage guide, installation instructions and additional figures are available for download at http://www.charite.de/sysbio/people/koenig/software/cysbml/.
|
|
| 2012 |

|
Quantifying the Contribution of the Liver to the Homeostasis of Plasma Glucose: A Detailed Kinetic Model of Hepatic Glucose Metabolism Integrated with the Hormonal Control by Insulin, Glucagon and Epinephrine
König M., Bulik S. and Holzhütter HG.
PLoS Comput Biol. 2012 Jun;8(6):e1002577. Epub 2012 Jun 21.. doi:10.1371/journal.pcbi.1002577. pmid:22761565
Despite the crucial role of the liver in glucose homeostasis, a detailed mathematical model of human hepatic glucose metabolism is lacking so far. Here we present a detailed kinetic model of glycolysis, gluconeogenesis and glycogen metabolism in human hepatocytes integrated with the hormonal control of these pathways by insulin, glucagon and epinephrine. Model simulations are in good agreement with experimental data on (i) the quantitative contributions of glycolysis, gluconeogenesis, and glycogen metabolism to hepatic glucose production and hepatic glucose utilization under varying physiological states. (ii) the time courses of postprandial glycogen storage as well as glycogen depletion in overnight fasting and short term fasting (iii) the switch from net hepatic glucose production under hypoglycemia to net hepatic glucose utilization under hyperglycemia essential for glucose homeostasis (iv) hormone perturbations of hepatic glucose metabolism. Response analysis reveals an extra high capacity of the liver to counteract changes of plasma glucose level below 5 mM (hypoglycemia) and above 7.5 mM (hyperglycemia). Our model may serve as an important module of a whole-body model of human glucose metabolism and as a valuable tool for understanding the role of the liver in glucose homeostasis under normal conditions and in diseases like diabetes or glycogen storage diseases.
|
|
| 2011 |

|
Enzymatic Features of the Glucose Metabolism in Tumor Cells.
Herling A, König M, Bulik S, Holzhütter HG.
FEBS J. 2011 Jul;278(14):2436-59. doi:10.1111/j.1742-4658.2011.08174.x. pmid:21564549
Many tumor types exhibit an impaired Pasteur effect, i.e. despite the presence of oxygen, glucose is consumed at an extraordinarily high rate compared with the tissue from which they originate - the so-called "Warburg effect". Glucose has to serve as the source for a diverse array of cellular functions, including energy production, synthesis of nucleotides and lipids, membrane synthesis and generation of redox equivalents for antioxidative defense. Tumor cells acquire specific enzyme-regulatory mechanisms to direct the main flux of glucose carbons to those pathways most urgently required under challenging external conditions such as varying substrate availability, presence of anti-cancer drugs or different phases of the cell cycle. In this review we summarize the currently available information on tumor-specific expression, activity and kinetic properties of enzymes involved in the main pathways of glucose metabolism with due regard to the explanation of the regulatory basis and physiological significance of the Warburg effect. We conclude that, besides the expression level of the metabolic enzymes involved in the glucose metabolism of tumor cells, the unique tumor-specific pattern of isozymes and accompanying changes in the metabolic regulation below the translation level enable tumor cells to drain selfishly the blood glucose pool that non-transformed cells use as sparingly as possible.
|
|
| 2011 |



|
FluxViz - Cytoscape Plug-in for Vizualisation of Flux Distributions in Networks
König M. and Holzhütter HG.
Genome Informatics 2010, Vol.24, p.96-103. pmid:22081592
Motivation: Methods like FBA and kinetic modeling are widely used to calculate fluxes in metabolic networks. For the analysis and understanding of simulation results and experimentally measured fluxes visualization software within the network context is indispensable. Results: We present Flux Viz, an open-source Cytoscape plug-in for the visualization of flux distributions in molecular interaction networks. FluxViz supports (i) import of networks in a variety of formats (SBML, GML, XGMML, SIF, BioPAX, PSI-MI) (ii) import of flux distributions as CSV, Cytoscape attributes or VAL files (iii) limitation of views to flux carrying reactions (flux subnetwork) or network attributes like localization (iv) export of generated views (SVG, EPS, PDF, BMP, PNG). Though FluxViz was primarily developed as tool for the visualization of fluxes in metabolic networks and the analysis of simulation results from FASIMU, a flexible software for batch flux-balance computation in large metabolic networks, it is not limited to biochemical reaction networks and FBA but can be applied to the visualization of arbitrary fluxes in arbitrary graphs. Availability: The platform-independent program is an open-source project, freely available at http://sourceforge.net/projects/fluxvizplugin/ under GNU public license, including manual, tutorial and examples.
|
|
| 2010 |

|
HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology.
Gille C, Bölling C, Hoppe A, Bulik S, Hoffmann S, Hübner K, Karlstädt A, Ganeshan R, König M, Rother K, Weidlich M, Behre J, Holzhütter HG.
Mol Syst Biol. 2010 Sep 7;6:411. doi:10.1038/msb.2010.62. pmid:20823849
We present HepatoNet1, the first reconstruction of a comprehensive metabolic network of the human hepatocyte that is shown to accomplish a large canon of known metabolic liver functions. The network comprises 777 metabolites in six intracellular and two extracellular compartments and 2539 reactions, including 1466 transport reactions. It is based on the manual evaluation of >1500 original scientific research publications to warrant a high-quality evidence-based model. The final network is the result of an iterative process of data compilation and rigorous computational testing of network functionality by means of constraint-based modeling techniques. Taking the hepatic detoxification of ammonia as an example, we show how the availability of nutrients and oxygen may modulate the interplay of various metabolic pathways to allow an efficient response of the liver to perturbations of the homeostasis of blood compounds.
|
|